Gene Expression Profiles Associated with Radio-Responsiveness in Locally Advanced Rectal Cancer
- PMID: 34205090
- PMCID: PMC8226560
- DOI: 10.3390/biology10060500
Gene Expression Profiles Associated with Radio-Responsiveness in Locally Advanced Rectal Cancer
Abstract
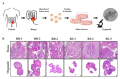
LARC patients were sorted according to their radio-responsiveness and patient-derived organoids were established from the respective cancer tissues. Expression profiles for each group were obtained using RNA-seq. Biological and bioinformatic analysis approaches were used in deciphering genes and pathways that participate in the radio-resistance of LARC. Thirty candidate genes encoding proteins involved in radio-responsiveness-related pathways, including the immune system, DNA repair and cell-cycle control, were identified. Interestingly, one of the candidate genes, cathepsin E (CTSE), exhibited differential methylation at the promoter region that was inversely correlated with the radio-resistance of patient-derived organoids, suggesting that methylation status could contribute to radio-responsiveness. On the basis of these results, we plan to pursue development of a gene chip for diagnosing the radio-responsiveness of LARC patients, with the hope that our efforts will ultimately improve the prognosis of LARC patients.
Keywords: DNA methylation; bio-marker; radiation therapy; radio-responsiveness; rectal cancer.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures







References
-
- Sauer R., Liersch T., Merkel S., Fietkau R., Hohenberger W., Hess C., Becker H., Raab H.R., Villanueva M.T., Witzigmann H., et al. Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: Results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J. Clin. Oncol. 2012;30:1926–1933. doi: 10.1200/JCO.2011.40.1836. - DOI - PubMed
-
- Kapiteijn E., Marijnen C.A., Nagtegaal I.D., Putter H., Steup W.H., Wiggers T., Rutten H.J., Pahlman L., Glimelius B., van Krieken J.H., et al. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer. N. Engl. J. Med. 2001;345:638–646. doi: 10.1056/NEJMoa010580. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

