FOXM1: A Multifunctional Oncoprotein and Emerging Therapeutic Target in Ovarian Cancer
- PMID: 34205406
- PMCID: PMC8235333
- DOI: 10.3390/cancers13123065
FOXM1: A Multifunctional Oncoprotein and Emerging Therapeutic Target in Ovarian Cancer
Abstract
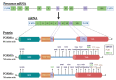
Forkhead box M1 (FOXM1) is a member of the conserved forkhead box (FOX) transcription factor family. Over the last two decades, FOXM1 has emerged as a multifunctional oncoprotein and a robust biomarker of poor prognosis in many human malignancies. In this review article, we address the current knowledge regarding the mechanisms of regulation and oncogenic functions of FOXM1, particularly in the context of ovarian cancer. FOXM1 and its associated oncogenic transcriptional signature are enriched in >85% of ovarian cancer cases and FOXM1 expression and activity can be enhanced by a plethora of genomic, transcriptional, post-transcriptional, and post-translational mechanisms. As a master transcriptional regulator, FOXM1 promotes critical oncogenic phenotypes in ovarian cancer, including: (1) cell proliferation, (2) invasion and metastasis, (3) chemotherapy resistance, (4) cancer stem cell (CSC) properties, (5) genomic instability, and (6) altered cellular metabolism. We additionally discuss the evidence for FOXM1 as a cancer biomarker, describe the rationale for FOXM1 as a cancer therapeutic target, and provide an overview of therapeutic strategies used to target FOXM1 for cancer treatment.
Keywords: FOXM1; forkhead box M1; high-grade serous ovarian cancer; oncogenes; oncoproteins; ovarian cancer; transcription factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Clevidence D.E., Overdier D.G., Tao W., Qian X., Pani L., Lai E., Costa R.H. Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc. Natl. Acad. Sci. USA. 1993;90:3948–3952. doi: 10.1073/pnas.90.9.3948. - DOI - PMC - PubMed
-
- Kaestner K.H., Knochel W., E Martinez D. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

