Development of a Bio-Layer Interferometry-Based Protease Assay Using HIV-1 Protease as a Model
- PMID: 34205716
- PMCID: PMC8235736
- DOI: 10.3390/v13061183
Development of a Bio-Layer Interferometry-Based Protease Assay Using HIV-1 Protease as a Model
Abstract
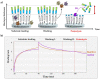
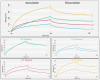
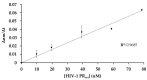
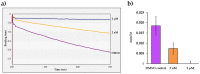
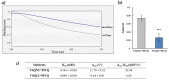
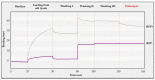
Proteolytic enzymes have great significance in medicine and the pharmaceutical industry and are applied in multiple fields of life sciences. Therefore, cost-efficient, reliable and sensitive real-time monitoring methods are highly desirable to measure protease activity. In this paper, we describe the development of a new experimental approach for investigation of proteolytic enzymes. The method was designed by the combination of recombinant fusion protein substrates and bio-layer interferometry (BLI). The protease (PR) of human immunodeficiency virus type 1 (HIV-1) was applied as model enzyme to set up and test the method. The principle of the assay is that the recombinant protein substrates immobilized to the surface of biosensor are specifically cleaved by the PR, and the substrate processing can be followed by measuring change in the layer thickness by optical measurement. We successfully used this method to detect the HIV-1 PR activity in real time, and the initial rate of the signal decrease was found to be proportional to the enzyme activity. Substrates representing wild-type and modified cleavage sites were designed to study HIV-1 PR's specificity, and the BLI-based measurements showed differential cleavage efficiency of the substrates, which was proven by enzyme kinetic measurements. We applied this BLI-based assay to experimentally confirm the existence of extended binding sites at the surface of HIV-1 PR. We found the measurements may be performed using lysates of cells expressing the fusion protein, without primary purification of the substrate. The designed BLI-based protease assay is high-throughput-compatible and enables real-time and small-volume measurements, thus providing a new and versatile approach to study proteolytic enzymes.
Keywords: BLItz; HIV-1; bio-layer interferometry; human immunodeficiency virus; protease; protease assay; recombinant fluorescent protein substrate.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

