Evaluation of a Novel CLIA Monotest Assay for the Detection of Anti-Hepatitis E Virus-IgG and IgM: A Retrospective Comparison with a Line Blot and an ELISA
- PMID: 34206114
- PMCID: PMC8228023
- DOI: 10.3390/pathogens10060689
Evaluation of a Novel CLIA Monotest Assay for the Detection of Anti-Hepatitis E Virus-IgG and IgM: A Retrospective Comparison with a Line Blot and an ELISA
Abstract
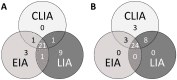
Despite the increasing relevance of Hepatitis E, an emerging disease endemic in developing and with increasing numbers of sporadic cases in industrialized countries, commercial tests are mainly based on batch oriented serological assays. In this retrospective study, we compared a line immunoassay (LIA; recomLine HEV, Mikrogen) and an ELISA (EIA; Anti-Hepatitis E Virus ELISA, Euroimmun) with a novel chemoluminescence immunoassay in a monotest format (CLIA; Hepatitis E VirClia, Vircell). Twenty sera of PCR proven cases of hepatitis E and 68 blood samples serologically pre-characterized were included. Applying the WHO reference standard, the CLIA demonstrated the highest analytical sensitivity for IgG and IgM. The combinations of CLIA/EIA (IgG and IgM) and CLIA/LIA (IgG) measurements showed substantial correlation. Compared to overall antibody detection (seropositivity in ≥2 assays), CLIA correlation was excellent, outperforming LIA (IgM) and EIA (IgG and IgM). Minor IgM cross reactivity in samples of patients with acute EBV infection was observed in all three assays. The CLIA showed good performance in diagnostic samples compared to established LIA and EIA assays. Due to its ready-to-use monotest format, the CLIA allows simple, time- and cost-effective handling of single samples. These qualities make the assay suitable for diagnostics, especially in the emergency setting and for low-throughput laboratories.
Keywords: CLIA; ELISA; HEV; LIA; antibody; hepatitis E virus; monotest; serology.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Assessment of the Diagnostic Performance of Fully Automated Hepatitis E Virus (HEV) Antibody Tests.Diagnostics (Basel). 2024 Mar 12;14(6):602. doi: 10.3390/diagnostics14060602. Diagnostics (Basel). 2024. PMID: 38535023 Free PMC article.
-
Monitoring of Anti-Hepatitis E Virus Antibody Seroconversion in Asymptomatically Infected Blood Donors: Systematic Comparison of Nine Commercial Anti-HEV IgM and IgG Assays.Viruses. 2016 Aug 22;8(8):232. doi: 10.3390/v8080232. Viruses. 2016. PMID: 27556482 Free PMC article.
-
Comparison of an Enzyme Linked-Immunosorbent Assay and a Chemiluminescent Immunoassay with an Immunofluorescence Assay for Detection of Phase II IgM and IgG Antibodies to Coxiella burnetii.Microorganisms. 2024 Mar 11;12(3):552. doi: 10.3390/microorganisms12030552. Microorganisms. 2024. PMID: 38543605 Free PMC article.
-
Comparative evaluation of chemiluminescent immunoassay and enzyme-linked immunosorbent assays for the diagnosis of West Nile virus infections.APMIS. 2022 Apr;130(4):215-220. doi: 10.1111/apm.13207. Epub 2022 Mar 1. APMIS. 2022. PMID: 35060204
-
Diagnostic performance of selected commercial HEV IgM and IgG ELISAs for immunocompromised and immunocompetent patients.J Clin Virol. 2013 Dec;58(4):629-34. doi: 10.1016/j.jcv.2013.10.010. Epub 2013 Oct 17. J Clin Virol. 2013. PMID: 24210958
Cited by
-
Development and Evaluation of an Immunochromatographic Strip and a Magnetic Chemiluminescence Immunoassay for Detection of Porcine Circovirus Type 2 Antigen.Vet Sci. 2025 Jan 9;12(1):40. doi: 10.3390/vetsci12010040. Vet Sci. 2025. PMID: 39852915 Free PMC article.
-
Assessment of the Diagnostic Performance of Fully Automated Hepatitis E Virus (HEV) Antibody Tests.Diagnostics (Basel). 2024 Mar 12;14(6):602. doi: 10.3390/diagnostics14060602. Diagnostics (Basel). 2024. PMID: 38535023 Free PMC article.
-
Development and Optimization of an Enzyme Immunoassay to Detect Serum Antibodies against the Hepatitis E Virus in Pigs, Using Plant-Derived ORF2 Recombinant Protein.Vaccines (Basel). 2021 Sep 6;9(9):991. doi: 10.3390/vaccines9090991. Vaccines (Basel). 2021. PMID: 34579228 Free PMC article.
-
Performance Comparison of Four Hepatitis E Antibodies Detection Methods.Microorganisms. 2024 Sep 11;12(9):1875. doi: 10.3390/microorganisms12091875. Microorganisms. 2024. PMID: 39338549 Free PMC article.
-
Diagnostic Performance of an Automated System for Assaying Anti-Hepatitis E Virus Immunoglobulins M and G Compared with a Conventional Microplate Assay.Viruses. 2022 May 17;14(5):1065. doi: 10.3390/v14051065. Viruses. 2022. PMID: 35632806 Free PMC article.
References
-
- Global Burden of Disease Study Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–1602. doi: 10.1016/S0140-6736(16)31678-6. - DOI - PMC - PubMed
-
- Pallerla S.R., Harms D., Johne R., Todt D., Steinmann E., Schemmerer M., Wenzel J.J., Hofmann J., Shih J.W.K., Wedemeyer H., et al. Hepatitis E Virus Infection: Circulation, Molecular Epidemiology, and Impact on Global Health. Pathogens. 2020;9:856. doi: 10.3390/pathogens9100856. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

