Biosensors Designed for Clinical Applications
- PMID: 34206405
- PMCID: PMC8301448
- DOI: 10.3390/biomedicines9070702
Biosensors Designed for Clinical Applications
Abstract
Emerging and validated biomarkers promise to revolutionize clinical practice, shifting the emphasis away from the management of chronic disease towards prevention, early diagnosis and early intervention. The challenge of detecting these low abundance protein and nucleic acid biomarkers within the clinical context demands the development of highly sensitive, even single molecule, assays that are also capable of selectively measuring a small number of defined analytes in complex samples such as whole blood, interstitial fluid, saliva or urine. Success relies on significant innovations in nanomaterials, bioreceptor engineering, transduction strategies and microfluidics. Primarily using examples from our work, this article discusses some recent advance in the selective and sensitive detection of disease biomarkers, highlights key innovations in sensor materials and identifies issues and challenges that need to be carefully considered especially for researchers entering the field.
Keywords: biomarkers; cancer; cardiovascular and neurological diseases; clinical analysis; electrochemical sensors; electrochemiluminescence; epilepsy; microfluidics; point of care.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






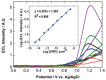
References
-
- [(accessed on 19 March 2021)]; Available online: https://jamaicahospital.org/newsletter/history-of-thermometers/
-
- Henning T.P., Cunningham D.D. Biosensors for Personal Diabetes Management. In: Ramsey G., editor. Commercial Biosensors: Applications to Clinical, Bioprocess, and Environmental Samples. John Wiley & Sons; New York, NY, USA: 1998. pp. 3–46.
-
- Forster R.J., Cumba L.R. Bioelectronics and Medical Devices. Woodhead Publishing; Cambridge, UK: 2019. Optimizing Glucose Sensing for Diabetes Monitoring; pp. 765–778.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

