Discovery of New Pyrazolopyridine, Furopyridine, and Pyridine Derivatives as CDK2 Inhibitors: Design, Synthesis, Docking Studies, and Anti-Proliferative Activity
- PMID: 34206976
- PMCID: PMC8272136
- DOI: 10.3390/molecules26133923
Discovery of New Pyrazolopyridine, Furopyridine, and Pyridine Derivatives as CDK2 Inhibitors: Design, Synthesis, Docking Studies, and Anti-Proliferative Activity
Abstract
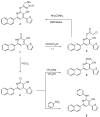
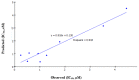
New pyridine, pyrazoloyridine, and furopyridine derivatives substituted with naphthyl and thienyl moieties were designed and synthesized starting from 6-(naphthalen-2-yl)-2-oxo-4-(thiophen-2-yl)-1,2-dihydropyridine-3-carbonitrile (1). The chloro, methoxy, cholroacetoxy, imidazolyl, azide, and arylamino derivatives were prepared to obtain the pyridine--C2 functionalized derivatives. The derived pyrazolpyridine-N-glycosides were synthesized via heterocyclization of the C2-thioxopyridine derivative followed by glycosylation using glucose and galactose. The furopyridine derivative 14 and the tricyclic pyrido[3',2':4,5]furo[3,2-d]pyrimidine 15 were prepared via heterocyclization of the ester derivative followed by a reaction with formamide. The newly synthesized compounds were evaluated for their ability to in vitro inhibit the CDK2 enzyme. In addition, the cytotoxicity of the compounds was tested against four different human cancer cell lines (HCT-116, MCF-7, HepG2, and A549). The CDK2/cyclin A2 enzyme inhibitory results revealed that pyridone 1, 2-chloro-6-(naphthalen-2-yl)-4-(thiophen-2-yl)nicotinonitrile (4), 6-(naphthalen-2-yl)-4-(thiophen-2-yl)-1H-pyrazolo[3,4-b]pyridin-3-amine (8), S-(3-cyano-6-(naphthaen-2-yl)-4-(thiophen-2-yl)pyridin-2-yl) 2-chloroethanethioate (11), and ethyl 3-amino-6-(naphthalen-2-yl)-4-(thiophen-2-yl)furo[2,3-b]pyridine-2-carboxylate (14) are among the most active inhibitors with IC50 values of 0.57, 0.24, 0.65, 0.50, and 0.93 µM, respectively, compared to roscovitine (IC50 0.394 μM). Most compounds showed significant inhibition on different human cancer cell lines (HCT-116, MCF-7, HepG2, and A549) with IC50 ranges of 31.3-49.0, 19.3-55.5, 22.7-44.8, and 36.8-70.7 μM, respectively compared to doxorubicin (IC50 40.0, 64.8, 24.7 and 58.1 µM, respectively). Furthermore, a molecular docking study suggests that most of the target compounds have a similar binding mode as a reference compound in the active site of the CDK2 enzyme. The structural requirements controlling the CDK2 inhibitory activity were determined through the generation of a statistically significant 2D-QSAR model.
Keywords: CDK2; HepG2; anticancer; docking; imidazole; pyrazolo[3,4-b]pyridine; pyridine.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


















References
-
- Alghassimou D., Claude P. The serine/threonine kinases that control cell cycle progression as therapeutic targets. Bull. Cancer. 2011;98:1335–1345. - PubMed
-
- Chen J., Wang X., Zhang J.Z., Zhu T. effect of substitution in different positions of aminothiazole hing-binding scaffolds on inhibitor-CDK2 association probed by interaction entropy method. ACS Omega. 2018;3:18052–18064. doi: 10.1021/acsomega.8b02354. - DOI
-
- Kontopidis G., McInnes C., Pandalaneni S.R., McNae I., Gibson D., Mezna M., Thomas M., Wood G., Wang S., Walkinshaw M.D., et al. Differential binding of inhibitors to active and inactive CDK2 provides insights for drug design. Chem. Biol. 2006;13:201–211. doi: 10.1016/j.chembiol.2005.11.011. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

