Human Defensins Inhibit SARS-CoV-2 Infection by Blocking Viral Entry
- PMID: 34206990
- PMCID: PMC8310277
- DOI: 10.3390/v13071246
Human Defensins Inhibit SARS-CoV-2 Infection by Blocking Viral Entry
Abstract
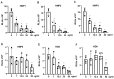
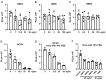
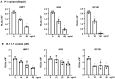
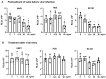
Innate immunity during acute infection plays a critical role in the disease severity of severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), and is likely to contribute to COVID-19 disease outcomes. Defensins are highly abundant innate immune factors in neutrophils and epithelial cells, including intestinal Paneth cells, and exhibit antimicrobial and immune-modulatory activities. In this study, we investigated the effects of human α- and β-defensins and RC101, a θ-defensin analog, on SARS-CoV-2 infection. We found that human neutrophil peptides (HNPs) 1-3, human defensin (HD) 5 and RC101 exhibited potent antiviral activity against pseudotyped viruses expressing SARS-CoV-2 spike proteins. HNP4 and HD6 had weak anti-SARS-CoV-2 activity, whereas human β-defensins (HBD2, HBD5 and HBD6) had no effect. HNP1, HD5 and RC101 also inhibited infection by replication-competent SARS-CoV-2 viruses and SARS-CoV-2 variants. Pretreatment of cells with HNP1, HD5 or RC101 provided some protection against viral infection. These defensins did not have an effect when provided post-infection, indicating their effect was directed towards viral entry. Indeed, HNP1 inhibited viral fusion but not the binding of the spike receptor-binding domain to hACE2. The anti-SARS-CoV-2 effect of defensins was influenced by the structure of the peptides, as linear unstructured forms of HNP1 and HD5 lost their antiviral function. Pro-HD5, the precursor of HD5, did not block infection by SARS-CoV-2. High virus titers overcame the effect of low levels of HNP1, indicating that defensins act on the virion. HNP1, HD5 and RC101 also blocked viral infection of intestinal and lung epithelial cells. The protective effects of defensins reported here suggest that they may be useful additives to the antivirus arsenal and should be thoroughly studied.
Keywords: SARS-CoV-2; antimicrobial peptides; defensins.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures


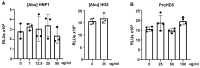
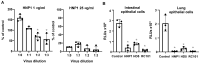
References
-
- [(accessed on 23 June 2021)]; Available online: https://coronavirus.jhu.edu/map.html.
-
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. - DOI - PMC - PubMed
-
- Channappanavar R., Fehr A.R., Zheng J., Wohlford-Lenane C., Abrahante J.E., Mack M., Sompallae R., McCray P.B., Jr., Meyerholz D.K., Perlman S. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Investig. 2019;129:3625–3639. doi: 10.1172/JCI126363. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

