Expression Profiling and Bioinformatics Analysis of CircRNA in Mice Brain Infected with Rabies Virus
- PMID: 34207166
- PMCID: PMC8234020
- DOI: 10.3390/ijms22126537
Expression Profiling and Bioinformatics Analysis of CircRNA in Mice Brain Infected with Rabies Virus
Abstract
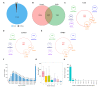
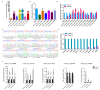
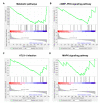
Rabies virus (RABV) induces acute, fatal encephalitis in mammals including humans. The circRNAs are important in virus infection process, but whether circRNAs regulated RABV infection remains largely unknown. Here, mice brain with or without the RABV CVS-11 strain were subjected to RNA sequencing and a total of 30,985 circRNAs were obtained. Among these, 9021 candidates were shared in both groups, and 14,610 and 7354 circRNAs were expressed specifically to the control and experimental groups, indicating that certain circRNAs were specifically inhibited or induced on RABV infection. The circRNAs mainly derived from coding exons. In total, 636 circRNAs were differentially expressed in RABV infection, of which 426 significantly upregulated and 210 significantly downregulated (p < 0.05 and fold change ≥2). The expression of randomly selected 6 upregulated and 6 downregulated circRNAs was tested by RT-qPCR, and the expression trend of the 11 out of 12 circRNAs was consistent in RT- qPCR and RNA-seq analysis. Rnase R-resistant assay and Sanger sequencing were conducted to verify the circularity of circRNAs. GO analysis demonstrated that source genes of all differentially regulated circRNAs were mainly related to cell plasticity and synapse function. Both KEGG and GSEA analysis revealed that these source genes were engaged in the cGMP-PKG and MAPK signaling pathway, and HTLV-I infection. Also, pathways related to glucose metabolism and synaptic functions were enriched in KEGG analysis. The circRNA-miRNA-mRNA network was built with 25 of 636 differentially expressed circRNAs, 264 mRNAs involved in RABV infection, and 29 miRNAs. Several miRNAs and many mRNAs in the network were reported to be related to viral infection and the immune response, suggesting that circRNAs could regulate RABV infection via interacting with miRNAs and mRNAs. Taken together, this study first characterized the transcriptomic pattern of circRNAs, and signaling pathways and function that circRNAs are involved in, which may indicate directions for further research to understand mechanisms of RABV pathogenesis.
Keywords: ceRNA network; circRNAs; enrichment analysis; mice brain; rabies virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Rieder M., Brzozka K., Pfaller C.K., Cox J.H., Stitz L., Conzelmann K.K. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprotein: Inhibition of interferon regulatory factor 3 activation is important for pathogenicity. J. Virol. 2011;85:842–852. doi: 10.1128/JVI.01427-10. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

