p53/p73 Protein Network in Colorectal Cancer and Other Human Malignancies
- PMID: 34207603
- PMCID: PMC8227208
- DOI: 10.3390/cancers13122885
p53/p73 Protein Network in Colorectal Cancer and Other Human Malignancies
Abstract
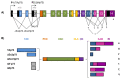
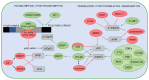
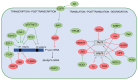
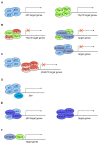
The p53 tumor suppressor protein is crucial for cell growth control and the maintenance of genomic stability. Later discovered, p63 and p73 share structural and functional similarity with p53. To understand the p53 pathways more profoundly, all family members should be considered. Each family member possesses two promoters and alternative translation initiation sites, and they undergo alternative splicing, generating multiple isoforms. The resulting isoforms have important roles in carcinogenesis, while their expression is dysregulated in several human tumors including colorectal carcinoma, which makes them potential targets in cancer treatment. Their activities arise, at least in part, from the ability to form tetramers that bind to specific DNA sequences and activate the transcription of target genes. In this review, we summarize the current understanding of the biological activities and regulation of the p53/p73 isoforms, highlighting their role in colorectal tumorigenesis. The analysis of the expression patterns of the p53/p73 isoforms in human cancers provides an important step in the improvement of cancer therapy. Furthermore, the interactions among the p53 family members which could modulate normal functions of the canonical p53 in tumor tissue are described. Lastly, we emphasize the importance of clinical studies to assess the significance of combining the deregulation of different members of the p53 family to define the outcome of the disease.
Keywords: colorectal cancer; isoform crosstalk; p53 family; p53 isoforms; p73 isoforms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Kaghad M., Bonnet H., Yang A., Creancier L., Biscan J.C., Valent A., Minty A., Chalon P., Lelias J.M., Dumont X., et al. Monoallelically Expressed Gene Related to P53 at 1p36, a Region Frequently Deleted in Neuroblastoma and Other Human Cancers. Cell. 1997;90:809–819. doi: 10.1016/S0092-8674(00)80540-1. - DOI - PubMed
-
- Yang A., Kaghad M., Wang Y., Gillett E., Fleming M.D., Dötsch V., Andrews N.C., Caput D., McKeon F. P63, a P53 Homolog at 3q27-29, Encodes Multiple Products with Transactivating, Death-Inducing, and Dominant-Negative Activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/S1097-2765(00)80275-0. - DOI - PubMed
-
- Moll U.M., Slade N. P63 and P73: Roles in Development and Tumor Formation. Mol. Cancer Res. 2004;2:371–386. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

