Assessment of Antidiabetic Activity of the Shikonin by Allosteric Inhibition of Protein-Tyrosine Phosphatase 1B (PTP1B) Using State of Art: An In Silico and In Vitro Tactics
- PMID: 34208908
- PMCID: PMC8271486
- DOI: 10.3390/molecules26133996
Assessment of Antidiabetic Activity of the Shikonin by Allosteric Inhibition of Protein-Tyrosine Phosphatase 1B (PTP1B) Using State of Art: An In Silico and In Vitro Tactics
Abstract
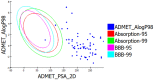
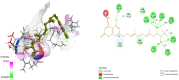
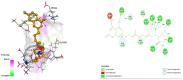
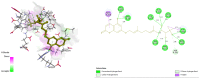
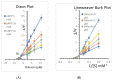
Diabetes mellitus is a multifactorial disease that affects both developing and developed countries and is a major public health concern. Many synthetic drugs are available in the market, which counteracts the associated pathologies. However, due to the propensity of side effects, there is an unmet need for the investigation of safe and effective drugs. This research aims to find a novel phytoconstituent having diminished action on blood glucose levels with the least side effects. Shikonin is a naturally occurring naphthoquinone dying pigment obtained by the roots of the Boraginaceae family. Besides its use as pigments, it can be used as an antimicrobial, anti-inflammatory, and anti-tumor agent. This research aimed to hypothesize the physicochemical and phytochemical properties of Shikonin's in silico interaction with protein tyrosine phosphate 1B, as well as it's in vitro studies, in order to determine its potential anti-diabetic impact. To do so, molecular docking experiments with target proteins were conducted to assess their anti-diabetic ability. Analyzing associations with corresponding amino acids revealed the significant molecular interactions between Shikonin and diabetes-related target proteins. In silico pharmacokinetics and toxicity profile of Shikonin using ADMET Descriptor, Toxicity Prediction, and Calculate Molecular Properties tools from Biovia Discovery Studio v4.5. Filter by Lipinski and Veber Rule's module from Biovia Discovery Studio v4.5 was applied to assess the drug-likeness of Shikonin. The in vitro studies exposed that Shikonin shows an inhibitory potential against the PTP1B with an IC50 value of 15.51 µM. The kinetics studies revealed that it has a competitive inhibitory effect (Ki = 7.5 M) on the enzyme system, which could be useful in the production of preventive and therapeutic agents. The findings of this research suggested that the Shikonin could be used as an anti-diabetic agent and can be used as a novel source for drug delivery.
Keywords: diabetes mellitus; hypoglycemic; molecular docking; protein-tyrosine phosphatase; shikonin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




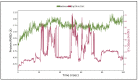
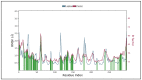
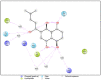
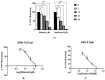
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

