Mutation Profiles, Glycosylation Site Distribution and Codon Usage Bias of Human Papillomavirus Type 16
- PMID: 34209097
- PMCID: PMC8310365
- DOI: 10.3390/v13071281
Mutation Profiles, Glycosylation Site Distribution and Codon Usage Bias of Human Papillomavirus Type 16
Abstract
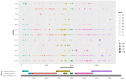
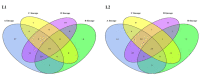
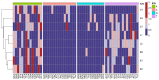
Human papillomavirus type 16 (HPV16) is the most prevalent HPV type causing cervical cancers. Herein, using 1597 full genomes, we systemically investigated the mutation profiles, surface protein glycosylation sites and the codon usage bias (CUB) of HPV16 from different lineages and sublineages. Multiple lineage- or sublineage-conserved mutation sites were identified. Glycosylation analysis showed that HPV16 lineage D contained the highest number of different glycosylation sites from lineage A in both L1 and L2 capsid proteins, which might lead to their antigenic distances between the two lineages. CUB analysis showed that the HPV16 open reading frames (ORFs) preferred codons ending with A/T. The CUB of HPV16 ORFs was mainly affected by natural selection except for E1, E5 and L2. HPV16 only shared some of the preferred codons with humans, which might help reduce competition in translational resources. These findings increase our understanding of the heterogeneity between HPV16 lineages and sublineages, and the adaptation mechanism of HPV in human cells. In summary, this study might facilitate HPV classification and improve vaccine development and application.
Keywords: HPV16; codon usage bias; glycosylation; lineage and sublineage; mutation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Comprehensive Assessment of the Antigenic Impact of Human Papillomavirus Lineage Variation on Recognition by Neutralizing Monoclonal Antibodies Raised against Lineage A Major Capsid Proteins of Vaccine-Related Genotypes.J Virol. 2020 Nov 23;94(24):e01236-20. doi: 10.1128/JVI.01236-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32967963 Free PMC article.
-
Differential expression of HPV16 L2 gene in cervical cancers harboring episomal HPV16 genomes: influence of synonymous and non-coding region variations.PLoS One. 2013 Jun 6;8(6):e65647. doi: 10.1371/journal.pone.0065647. Print 2013. PLoS One. 2013. PMID: 23762404 Free PMC article.
-
Genetic signatures for lineage/sublineage classification of HPV16, 18, 52 and 58 variants.Virology. 2021 Jan 15;553:62-69. doi: 10.1016/j.virol.2020.11.003. Epub 2020 Nov 16. Virology. 2021. PMID: 33238224
-
Methylation at 3'LCR of HPV16 can be affected by patient age and disruption of E1 or E2 genes.Virus Res. 2017 Mar 15;232:48-53. doi: 10.1016/j.virusres.2017.01.022. Epub 2017 Jan 29. Virus Res. 2017. PMID: 28143725
-
Recent Advances in Our Understanding of the Infectious Entry Pathway of Human Papillomavirus Type 16.Microorganisms. 2021 Oct 1;9(10):2076. doi: 10.3390/microorganisms9102076. Microorganisms. 2021. PMID: 34683397 Free PMC article. Review.
Cited by
-
Differences in HPV-specific antibody Fc-effector functions following Gardasil® and Cervarix® vaccination.NPJ Vaccines. 2023 Mar 15;8(1):39. doi: 10.1038/s41541-023-00628-8. NPJ Vaccines. 2023. PMID: 36922512 Free PMC article.
-
Distinguishing Genetic Drift from Selection in Papillomavirus Evolution.Viruses. 2023 Jul 26;15(8):1631. doi: 10.3390/v15081631. Viruses. 2023. PMID: 37631973 Free PMC article.
-
Comprehensive Analysis of Codon Usage Bias in Human Papillomavirus Type 51.Pol J Microbiol. 2024 Oct 28;73(4):455-465. doi: 10.33073/pjm-2024-036. eCollection 2024 Dec 1. Pol J Microbiol. 2024. PMID: 39465910 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

