Consequences and Resolution of Transcription-Replication Conflicts
- PMID: 34209204
- PMCID: PMC8303131
- DOI: 10.3390/life11070637
Consequences and Resolution of Transcription-Replication Conflicts
Abstract
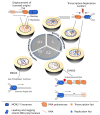
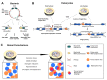
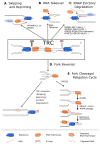
Transcription-replication conflicts occur when the two critical cellular machineries responsible for gene expression and genome duplication collide with each other on the same genomic location. Although both prokaryotic and eukaryotic cells have evolved multiple mechanisms to coordinate these processes on individual chromosomes, it is now clear that conflicts can arise due to aberrant transcription regulation and premature proliferation, leading to DNA replication stress and genomic instability. As both are considered hallmarks of aging and human diseases such as cancer, understanding the cellular consequences of conflicts is of paramount importance. In this article, we summarize our current knowledge on where and when collisions occur and how these encounters affect the genome and chromatin landscape of cells. Finally, we conclude with the different cellular pathways and multiple mechanisms that cells have put in place at conflict sites to ensure the resolution of conflicts and accurate genome duplication.
Keywords: G-MiDS; MIDAS; R-loops; chromatin; common fragile sites; early replicating fragile sites; fork reversal; genomic instability; replication stress; torsional stress; transcription–replication conflicts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Kurat C.F., Lambert J.P., van Dyk D., Tsui K., van Bakel H., Kaluarachchi S., Friesen H., Kainth P., Nislow C., Figeys D., et al. Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein. Genes Dev. 2011;25:2489–2501. doi: 10.1101/gad.173427.111. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

