Engraftment of Bacteria after Fecal Microbiota Transplantation Is Dependent on Both Frequency of Dosing and Duration of Preparative Antibiotic Regimen
- PMID: 34209573
- PMCID: PMC8306289
- DOI: 10.3390/microorganisms9071399
Engraftment of Bacteria after Fecal Microbiota Transplantation Is Dependent on Both Frequency of Dosing and Duration of Preparative Antibiotic Regimen
Abstract
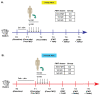
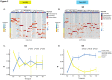
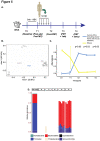
The gut microbiota has emerged as a key mediator of human physiology, and germ-free mice have been essential in demonstrating a role for the microbiome in disease. Preclinical models using conventional mice offer the advantage of working with a mature immune system. However, optimal protocols for fecal microbiota transplant (FMT) engraftment in conventional mice are yet to be established. Conventional BALB/c mice were randomized to receive 3-day (3d) or 3-week (3w) antibiotic (ABX) regimen in their drinking water followed by 1 or 5-daily FMTs from a human donor. Fecal samples were collected longitudinally and characterized using 16S ribosomal RNA (rRNA) sequencing. Semi-targeted metabolomic profiling of fecal samples was also done with liquid chromatography-mass spectrometry (LC-MS). Lastly, we sought to confirm our findings in BKS mice. Recovery of baseline diversity scores were greatest in the 3d groups, driven by re-emergence of mouse commensal microbiota, whereas the most resemblance to donor microbiota was seen in the 3w + 5-FMT group. Amplicon sequence variants (ASVs) that were linked to the input material (human ASVs) engrafted to a significantly greater extent when compared to mouse ASVs in the 3-week groups but not the 3-day groups. Lastly, comparison of metabolomic profiles revealed distinct functional profiles by ABX regimen. These results indicate successful model optimization and emphasize the importance of ABX duration and frequency of FMT dosing; the most stable and reliable colonization by donor ASVs was seen in the 3wk + 5-FMT group.
Keywords: antibiotics; conventional mice; engraftment; microbiome.
Conflict of interest statement
V.G. is an inventor on a US patent application (PCT/US17/53,717), submitted by The University of Texas MD Anderson Cancer Center, that covers methods to enhance checkpoint blockade therapy by the microbiome. V.G., E.A.D., M.F., M.G., P.W., C.C., S.H., B.R.S., T.S.C. are employed by, and own stock in AstraZeneca.
Figures


References
-
- Thomas S., Izard J., Walsh E., Batich K., Chongsathidkiet P., Clarke G., Sela D.A., Muller A.J., Mullin J.M., Albert K., et al. The host microbiome regulates and maintains human health: A primer and perspective for non-microbiologists. Cancer Res. 2017;77:1783–1812. doi: 10.1158/0008-5472.CAN-16-2929. - DOI - PMC - PubMed
-
- Davar D., Dzutsev A.K., McCulloch J.A., Rodrigues R.R., Chauvin J.M., Morrison R.M., Deblasio R.N., Menna C., Ding Q., Pagliano O., et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science. 2021;371:595–602. doi: 10.1126/science.abf3363. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

