Comparison of Hepatitis E Virus Sequences from Humans and Swine, the Netherlands, 1998-2015
- PMID: 34209729
- PMCID: PMC8310231
- DOI: 10.3390/v13071265
Comparison of Hepatitis E Virus Sequences from Humans and Swine, the Netherlands, 1998-2015
Abstract
Pigs are suspected to be a major source of zoonotic hepatitis E virus (HEV) infection in industrialized countries, but the transmission route(s) from pigs to humans are ill-defined. Sequence comparison of HEV isolates from pigs with those from blood donors and patients in 372 samples collected in the Netherlands in 1998 and 1999 and between 2008 and 2015 showed that all sequences were genotype 3 except for six patients (with travel history). Subgenotype 3c (gt3c) was the most common subtype. While the proportion of gt3c increased significantly between 1998 and 2008, it remained constant between 2008 and 2015. Among the few circulating HEV subtypes, there was no difference observed between the human and the pig isolates. Hepatitis E viruses in humans are very likely to originate from pigs, but it is unclear why HEV gt3c has become the predominant subtype in the Netherlands.
Keywords: hepatitis E; phylogenetic analysis; zoonotic infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
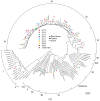
 ), patients (■) and pigs (
), patients (■) and pigs ( ). Phylogenetic analysis of the 242 bp ORF1 fragment and the 304 bp ORF2 fragment was performed in MEGA and visualized using FigTree. A selection of genotype 1–3 sequences with assigned (sub)genotype were used as reference sequences; the sequences from the proposed reference set are indicated with an asterisk (*) [4]. The circle around the tree indicates the subtypes; if no space was available subtypes were combined as indicated (e.g., 3h/i). The scale bar at the bottom right denotes evolutionary distance.
). Phylogenetic analysis of the 242 bp ORF1 fragment and the 304 bp ORF2 fragment was performed in MEGA and visualized using FigTree. A selection of genotype 1–3 sequences with assigned (sub)genotype were used as reference sequences; the sequences from the proposed reference set are indicated with an asterisk (*) [4]. The circle around the tree indicates the subtypes; if no space was available subtypes were combined as indicated (e.g., 3h/i). The scale bar at the bottom right denotes evolutionary distance.
 ), patients (■) and pigs (
), patients (■) and pigs ( ). Phylogenetic analysis of the 242 bp ORF1 fragment and the 304 bp ORF2 fragment was performed in MEGA and visualized using FigTree. A selection of genotype 1–3 sequences with assigned (sub)genotype were used as reference sequences; the sequences from the proposed reference set are indicated with an asterisk (*) [4]. The circle around the tree indicates the subtypes; if no space was available subtypes were combined as indicated (e.g., 3h/i). The scale bar at the bottom right denotes evolutionary distance.
). Phylogenetic analysis of the 242 bp ORF1 fragment and the 304 bp ORF2 fragment was performed in MEGA and visualized using FigTree. A selection of genotype 1–3 sequences with assigned (sub)genotype were used as reference sequences; the sequences from the proposed reference set are indicated with an asterisk (*) [4]. The circle around the tree indicates the subtypes; if no space was available subtypes were combined as indicated (e.g., 3h/i). The scale bar at the bottom right denotes evolutionary distance.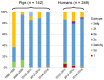
References
-
- Smith D.B., Simmonds P., International Committee on Taxonomy of Viruses Hepeviridae Study Group. Jameel S., Emerson S.U., Harrison T.J., Meng X.-J., Okamoto H., Van der Poel W.H.M., Purdy M.A. Consensus proposals for classification of the family Hepeviridae. Pt 10J. Gen. Virol. 2014;95:2223–2232. doi: 10.1099/vir.0.068429-0. - DOI - PMC - PubMed
-
- Lee G.H., Tan B.H., Teo E.C., Lim S.G., Dan Y.Y., Wee A., Aw P.P., Zhu Y., Hibberd M.L., Tan C.K., et al. Chronic Infection with Camelid Hepatitis E Virus in a Liver Transplant Recipient Who Regularly Consumes Camel Meat and Milk. Gastroenterology. 2016;150:355–357. doi: 10.1053/j.gastro.2015.10.048. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

