Effects of the Developmental Regulator BOLITA on the Plant Metabolome
- PMID: 34209960
- PMCID: PMC8305173
- DOI: 10.3390/genes12070995
Effects of the Developmental Regulator BOLITA on the Plant Metabolome
Abstract
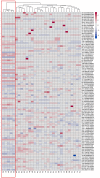
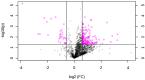
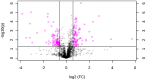
Transcription factors are important regulators of gene expression. They can orchestrate the activation or repression of hundreds or thousands of genes and control diverse processes in a coordinated way. This work explores the effect of a master regulator of plant development, BOLITA (BOL), in plant metabolism, with a special focus on specialized metabolism. For this, we used an Arabidopsis thaliana line in which the transcription factor activity can be induced. Fingerprinting metabolomic analyses of whole plantlets were performed at different times after induction. After 96 h, all induced replicas clustered as a single group, in contrast with all controls which did not cluster. Metabolomic analyses of shoot and root tissues enabled the putative identification of differentially accumulated metabolites in each tissue. Finally, the analysis of global gene expression in induced vs. non-induced root samples, together with enrichment analyses, allowed the identification of enriched metabolic pathways among the differentially expressed genes and accumulated metabolites after the induction. We concluded that the induction of BOL activity can modify the Arabidopsis metabolome. Future work should investigate whether its action is direct or indirect, and the implications of the metabolic changes for development regulation and bioprospection.
Keywords: developmental regulation; global expression analysis; glucosinolates; metabolic fingerprinting; phenylpropanoid pathway; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Roll-Hansen N. The Genotype Theory of Wilhelm Johannsen and its Relation to Plant Breeding and the Study of Evolution. Centaurus. 1979;22:201–235. doi: 10.1111/j.1600-0498.1979.tb00589.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

