Detection of Q129H Immune Escape Mutation in Apparently Healthy Hepatitis B Virus Carriers in Southwestern Nigeria
- PMID: 34210073
- PMCID: PMC8310067
- DOI: 10.3390/v13071273
Detection of Q129H Immune Escape Mutation in Apparently Healthy Hepatitis B Virus Carriers in Southwestern Nigeria
Abstract
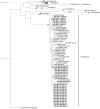
As the global effort to eradicate hepatitis B continues, immune escape mutations (IEMs) and drug resistance mutations (DRMs) affecting its diagnosis, treatment, and prevention are compromising this goal. However, knowledge about the prevalence and circulation of these mutations in Nigeria is scarce. Serum samples (n = 199) from apparently healthy prospective blood donors, pregnant women, and individuals presenting with fever in southwestern Nigeria were analyzed for the presence of IEMs and DRMs by means of nested PCR in the HBV S (HBs) and HBV polymerase (Pol) genes, followed by phylogenetic and mutational analyses. In total, 25.1% (n = 50/199) of samples were positive for HBV, as measured by PCR. In 41 samples (20.6%), both fragments could be amplified, whereas the HBs gene and the Pol gene fragment alone were detected in 0.5% (n = 1/199) and 4% (n = 8/199) of samples, respectively. Sequences were successfully obtained for all 42 HBs gene fragments but for only 31/49 Pol gene fragments (totaling 73 sequences from 44 individuals). All sequences were identified as HBV genotype E. IEMs were present in 18.2% (n = 8/44) of the sequences of HBV-positive individuals with available sequences. IEM Q129H was detected in eight out of the 44 (18.2%) HBV isolates sequenced in this study; however, no DRMs were observed. This study confirms the circulation of HBV IEMs and reports the presence of Q129H IEM for the first time in Nigeria. Intensified research on the dynamics of IEM is necessary in order to enhance the elimination of HBV.
Keywords: Nigeria; PCR; hepatitis; mutation; phylogeny; resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

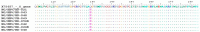
References
-
- WHO Hepatitis B. [(accessed on 2 October 2020)]; Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b.
-
- Harrison A., Lemey P., Hurles M., Moyes C., Horn S., Pryor J., Malani J., Supuri M., Masta A., Teriboriki B., et al. Genomic analysis of hepatitis B virus reveals antigen state and genotype as sources of evolutionary rate variation. Viruses. 2011;3:83–101. doi: 10.3390/v3020083. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

