Pathogenic SPTBN1 variants cause an autosomal dominant neurodevelopmental syndrome
- PMID: 34211179
- PMCID: PMC8273149
- DOI: 10.1038/s41588-021-00886-z
Pathogenic SPTBN1 variants cause an autosomal dominant neurodevelopmental syndrome
Abstract
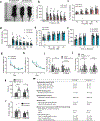
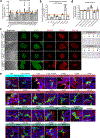
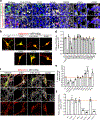
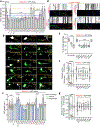
SPTBN1 encodes βII-spectrin, the ubiquitously expressed β-spectrin that forms micrometer-scale networks associated with plasma membranes. Mice deficient in neuronal βII-spectrin have defects in cortical organization, developmental delay and behavioral deficiencies. These phenotypes, while less severe, are observed in haploinsufficient animals, suggesting that individuals carrying heterozygous SPTBN1 variants may also show measurable compromise of neural development and function. Here we identify heterozygous SPTBN1 variants in 29 individuals with developmental, language and motor delays; mild to severe intellectual disability; autistic features; seizures; behavioral and movement abnormalities; hypotonia; and variable dysmorphic facial features. We show that these SPTBN1 variants lead to effects that affect βII-spectrin stability, disrupt binding to key molecular partners, and disturb cytoskeleton organization and dynamics. Our studies define SPTBN1 variants as the genetic basis of a neurodevelopmental syndrome, expand the set of spectrinopathies affecting the brain and underscore the critical role of βII-spectrin in the central nervous system.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare the following competing interests: E.T., R.E.P., Y.S., E.A.N., and A.B. are employees of GeneDx, Inc.
Figures








References
-
- Bennett V & Lorenzo DN Spectrin- and ankyrin-based membrane domains and the evolution of vertebrates. Curr. Top. Membr 72, 1–37 (2013). - PubMed
-
- Bennett V & Lorenzo DN An adaptable spectrin/ankyrin-based mechanism for long-range organization of plasma membranes in vertebrate tissues. Curr. Top. Membr 77, 143–184 (2016). - PubMed
-
- Lorenzo DN Cargo hold and delivery: Ankyrins, spectrins, and their functional patterning of neurons. Cytoskeleton 77, 129–148 (2020). - PubMed
-
- Ikeda Y et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat. Genet 38, 184–190 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

