Bioinformatics Analysis of ceRNA Network Related to Polycystic Ovarian Syndrome
- PMID: 34211581
- PMCID: PMC8208863
- DOI: 10.1155/2021/9988347
Bioinformatics Analysis of ceRNA Network Related to Polycystic Ovarian Syndrome
Abstract
Introduction: Polycystic ovary syndrome (PCOS) is caused by the hormonal environment in utero, abnormal metabolism, and genetics, and it is common in women of childbearing age. A large number of studies have reported that lncRNA is important to the biological process of cancer and can be used as a potential prognostic biomarker. Thus, we studied lncRNAs' roles in PCOS in this article.
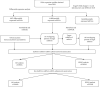
Methods: We obtained mRNAs', miRNAs', and lncRNAs' expression profiles in PCOS specimens and normal specimens from the National Biotechnology Information Gene Expression Comprehensive Center database. The EdgeR software package is used to distinguish the differentially expressed lncRNAs, miRNAs, and mRNAs. Functional enrichment analysis was carried out by the clusterProfiler R Package, and the lncRNA-miRNA-mRNA interaction ceRNA network was built in Cytoscape plug-in BiNGO and Database for Annotation, Visualization, and Integration Discovery (DAVID), respectively.
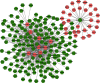
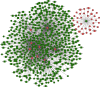
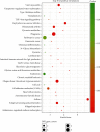
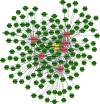
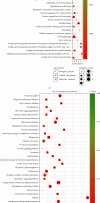
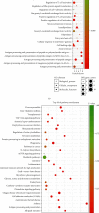
Results: We distinguished differentially expressed RNAs, including 1087 lncRNAs, 14 miRNAs, and 566 mRNAs in PCOS. Among them, 410 lncRNAs, 11 miRNAs, and 185 mRNAs were contained in the ceRNA regulatory network. The outcomes from Gene Ontology (GO) analysis showed that the differentially expressed mRNAs (DEMs) were mainly enriched in response to the maternal process involved in female pregnancy, morphogenesis of embryonic epithelium, and the intracellular steroid hormone receptor signaling pathway. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis data showed that DEMs were primarily enriched in pathways related to the TGF-β signaling pathway, Type I diabetes mellitus, and glycolysis/gluconeogenesis. In addition, we chose NONHSAT123397, ENST00000564619, and NONHSAT077997 as key lncRNAs due to their high bearing on PCOS.
Conclusion: ceRNA networks play an important role in PCOS. The research indicated that specific lncRNAs were related to PCOS development. NONHSAT123397, ENST00000564619, and NONHSAT077997 could be regarded as potential diagnostic mechanisms and biomarkers for PCOS. This discovery might provide more effective and more novel insights into the mechanisms of PCOS worthy of further exploration.
Copyright © 2021 Yuanqi Li and Yong Tan.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

