Identification of a Seven-lncRNA-mRNA Signature for Recurrence and Prognostic Prediction in Relapsed Acute Lymphoblastic Leukemia Based on WGCNA and LASSO Analyses
- PMID: 34211824
- PMCID: PMC8208884
- DOI: 10.1155/2021/6692022
Identification of a Seven-lncRNA-mRNA Signature for Recurrence and Prognostic Prediction in Relapsed Acute Lymphoblastic Leukemia Based on WGCNA and LASSO Analyses
Abstract
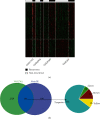
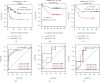
Abnormal expressions of long noncoding RNAs (lncRNAs) and protein-encoding messenger RNAs (mRNAs) are important for the development of childhood acute lymphoblastic leukemia (ALL). This study developed an lncRNA-mRNA integrated classifier for the prediction of recurrence and prognosis in relapsed childhood ALL by using several transcriptome data. Weighted gene coexpression network analysis revealed that green, turquoise, yellow, and brown modules were preserved across the TARGET, GSE60926, GSE28460, and GSE17703 datasets, and they were associated with clinical relapse and death status. A total of 184 genes in these four modules were differentially expressed between recurrence and nonrecurrence samples. Least absolute shrinkage and selection operator analysis showed that seven genes constructed a prognostic signature (including one lncRNA: LINC00652 and six mRNAs: INSL3, NIPAL2, REN, RIMS2, RPRM, and SNAP91). Kaplan-Meier curve analysis observed that patients in the high-risk group had a significantly shorter overall survival than those of the low-risk group. Receiver operating characteristic curve analysis demonstrated that this signature had high accuracy in predicting the 5-year overall survival and recurrence outcomes, respectively. LINC00652 may function by coexpressing with the above prognostic genes (INSL3, SNAP91, and REN) and lipid metabolism-related genes (MIA2, APOA1). Accordingly, this lncRNA-mRNA-based classifier may be clinically useful to predict the recurrence and prognosis for childhood ALL. These genes represent new targets to explain the mechanisms for ALL.
Copyright © 2021 Haiyan Qi et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Abdel Ghafar M. T., Gharib F., al-Ashmawy G. M., Mariah R. A. Serum high-temperature-required protein A2: a potential biomarker for the diagnosis of breast cancer. Gene Reports. 2020;20:p. 100706. doi: 10.1016/j.genrep.2020.100706. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

