nZVI Impacts Substrate Conversion and Microbiome Composition in Chain Elongation From D- and L-Lactate Substrates
- PMID: 34211964
- PMCID: PMC8239352
- DOI: 10.3389/fbioe.2021.666582
nZVI Impacts Substrate Conversion and Microbiome Composition in Chain Elongation From D- and L-Lactate Substrates
Abstract
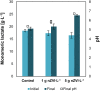
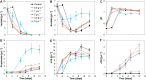
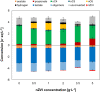
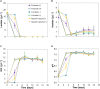
Medium-chain carboxylates (MCC) derived from biomass biorefining are attractive biochemicals to uncouple the production of a wide array of products from the use of non-renewable sources. Biological conversion of biomass-derived lactate during secondary fermentation can be steered to produce a variety of MCC through chain elongation. We explored the effects of zero-valent iron nanoparticles (nZVI) and lactate enantiomers on substrate consumption, product formation and microbiome composition in batch lactate-based chain elongation. In abiotic tests, nZVI supported chemical hydrolysis of lactate oligomers present in concentrated lactic acid. In fermentation experiments, nZVI created favorable conditions for either chain-elongating or propionate-producing microbiomes in a dose-dependent manner. Improved lactate conversion rates and n-caproate production were promoted at 0.5-2 g nZVI⋅L-1 while propionate formation became relevant at ≥ 3.5 g nZVI⋅L-1. Even-chain carboxylates (n-butyrate) were produced when using enantiopure and racemic lactate with lactate conversion rates increased in nZVI presence (1 g⋅L-1). Consumption of hydrogen and carbon dioxide was observed late in the incubations and correlated with acetate formation or substrate conversion to elongated products in the presence of nZVI. Lactate racemization was observed during chain elongation while isomerization to D-lactate was detected during propionate formation. Clostridium luticellarii, Caproiciproducens, and Ruminococcaceae related species were associated with n-valerate and n-caproate production while propionate was likely produced through the acrylate pathway by Clostridium novyi. The enrichment of different potential n-butyrate producers (Clostridium tyrobutyricum, Lachnospiraceae, Oscillibacter, Sedimentibacter) was affected by nZVI presence and concentrations. Possible theories and mechanisms underlying the effects of nZVI on substrate conversion and microbiome composition are discussed. An outlook is provided to integrate (bio)electrochemical systems to recycle (n)ZVI and provide an alternative reducing power agent as durable control method.
Keywords: carboxylates; lactate isomer metabolism; lactate isomerization; lactate racemization; microbial chain elongation; n-butyrate; n-caproate; zero-valent iron.
Copyright © 2021 Contreras-Dávila, Esveld, Buisman and Strik.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Akedo M., Cooney C. L., Sinskey A. J. (1983). Direct demonstration of lactate-acrylate interconversion in Clostridium propionicum. Nat. Biotechnol. 1 791–794.
-
- Andrews J. H., Harris R. F. (1986). “r- and K-Selection and Microbial Ecology,” in Advances in Microbial Ecology, ed. Marshall K. C. (Boston, MA: Springer; ), 99–147. 10.1007/978-1-4757-0611-6_3 - DOI
-
- Beck M. W., Mikryukov V. (2020). ggord: Ordination Plots with ggplot2 R package version 1.0. 10.5281/zenodo.842225 - DOI
LinkOut - more resources
Full Text Sources

