Site-Specific Steric Control of SARS-CoV-2 Spike Glycosylation
- PMID: 34213308
- PMCID: PMC8262170
- DOI: 10.1021/acs.biochem.1c00279
Site-Specific Steric Control of SARS-CoV-2 Spike Glycosylation
Abstract
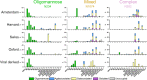
A central tenet in the design of vaccines is the display of native-like antigens in the elicitation of protective immunity. The abundance of N-linked glycans across the SARS-CoV-2 spike protein is a potential source of heterogeneity among the many different vaccine candidates under investigation. Here, we investigate the glycosylation of recombinant SARS-CoV-2 spike proteins from five different laboratories and compare them against S protein from infectious virus, cultured in Vero cells. We find patterns that are conserved across all samples, and this can be associated with site-specific stalling of glycan maturation that acts as a highly sensitive reporter of protein structure. Molecular dynamics simulations of a fully glycosylated spike support a model of steric restrictions that shape enzymatic processing of the glycans. These results suggest that recombinant spike-based SARS-CoV-2 immunogen glycosylation reproducibly recapitulates signatures of viral glycosylation.
Conflict of interest statement
The authors declare the following competing financial interest(s): ExcellGene sells purified trimeric spike protein preparations from CHO cells to commercial companies for internal research and for use in diagnostic applications.
Figures


Update of
-
Site-specific steric control of SARS-CoV-2 spike glycosylation.bioRxiv [Preprint]. 2021 Mar 9:2021.03.08.433764. doi: 10.1101/2021.03.08.433764. bioRxiv. 2021. Update in: Biochemistry. 2021 Jul 13;60(27):2153-2169. doi: 10.1021/acs.biochem.1c00279. PMID: 33758835 Free PMC article. Updated. Preprint.
References
-
- Brouwer P. J. M.; Caniels T. G.; van der Straten K.; Snitselaar J. L.; Aldon Y.; Bangaru S.; Torres J. L.; Okba N. M. A.; Claireaux M.; Kerster G.; Bentlage A. E. H.; van Haaren M. M.; Guerra D.; Burger J. A.; Schermer E. E.; Verheul K. D.; van der Velde N.; van der Kooi A.; van Schooten J.; van Breemen M. J.; Bijl T. P. L.; Sliepen K.; Aartse A.; Derking R.; Bontjer I.; Kootstra N. A.; Wiersinga W. J.; Vidarsson G.; Haagmans B. L.; Ward A. B.; de Bree G. J.; Sanders R. W.; van Gils M. J. (2020) Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650. 10.1126/science.abc5902. - DOI - PMC - PubMed
-
- Pinto D.; Park Y. J.; Beltramello M.; Walls A. C.; Tortorici M. A.; Bianchi S.; Jaconi S.; Culap K.; Zatta F.; De Marco A.; Peter A.; Guarino B.; Spreafico R.; Cameroni E.; Case J. B.; Chen R. E.; Havenar-Daughton C.; Snell G.; Telenti A.; Virgin H. W.; Lanzavecchia A.; Diamond M. S.; Fink K.; Veesler D.; Corti D. (2020) Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295. 10.1038/s41586-020-2349-y. - DOI - PubMed
-
- Liu L.; Wang P.; Nair M. S.; Yu J.; Rapp M.; Wang Q.; Luo Y.; Chan J. F. W.; Sahi V.; Figueroa A.; Guo X. V.; Cerutti G.; Bimela J.; Gorman J.; Zhou T.; Chen Z.; Yuen K. Y.; Kwong P. D.; Sodroski J. G.; Yin M. T.; Sheng Z.; Huang Y.; Shapiro L.; Ho D. D. (2020) Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456. 10.1038/s41586-020-2571-7. - DOI - PubMed
-
- Polack F. P.; Thomas S. J.; Kitchin N.; Absalon J.; Gurtman A.; Lockhart S.; Perez J. L.; Pérez Marc G.; Moreira E. D.; Zerbini C.; Bailey R.; Swanson K. A.; Roychoudhury S.; Koury K.; Li P.; Kalina W. V.; Cooper D.; Frenck R. W.; Hammitt L. L.; Türeci Ö.; Nell H.; Schaefer A.; Ünal S.; Tresnan D. B.; Mather S.; Dormitzer P. R.; Şahin U.; Jansen K. U.; Gruber W. C. (2020) Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 383, 2603–2615. 10.1056/NEJMoa2034577. - DOI - PMC - PubMed
-
- Baden L. R.; El Sahly H. M.; Essink B.; Kotloff K.; Frey S.; Novak R.; Diemert D.; Spector S. A.; Rouphael N.; Creech C. B.; McGettigan J.; Khetan S.; Segall N.; Solis J.; Brosz A.; Fierro C.; Schwartz H.; Neuzil K.; Corey L.; Gilbert P.; Janes H.; Follmann D.; Marovich M.; Mascola J.; Polakowski L.; Ledgerwood J.; Graham B. S.; Bennett H.; Pajon R.; Knightly C.; Leav B.; Deng W.; Zhou H.; Han S.; Ivarsson M.; Miller J.; Zaks T. (2021) Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 384, 403–416. 10.1056/NEJMoa2035389. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

