Evaluation of a Rapid Antigen Test To Detect SARS-CoV-2 Infection and Identify Potentially Infectious Individuals
- PMID: 34213977
- PMCID: PMC8373008
- DOI: 10.1128/JCM.00896-21
Evaluation of a Rapid Antigen Test To Detect SARS-CoV-2 Infection and Identify Potentially Infectious Individuals
Abstract
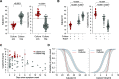
The identification and isolation of highly infectious SARS-CoV-2-infected individuals is an important public health strategy. Rapid antigen detection tests (RADT) are promising tools for large-scale screenings due to timely results and feasibility for on-site testing. Nonetheless, the diagnostic performance of RADT in detecting infectious individuals is not yet fully determined. In this study, RT-qPCR and virus culture of RT-qPCR-positive samples were used to evaluate and compare the performance of the Standard Q COVID-19 Ag test in detecting SARS-CoV-2-infected and possibly infectious individuals. To this end, two combined oro- and nasopharyngeal swabs were collected at a routine SARS-CoV-2 diagnostic center. A total of 2,028 samples were tested, and 118 virus cultures were inoculated. SARS-CoV-2 infection was detected in 210 samples by RT-qPCR, representing a positive rate of 10.36%. The Standard Q COVID-19 Ag test yielded a positive result in 92 (4.54%) samples resulting in an overall sensitivity and specificity of 42.86 and 99.89%, respectively. For adjusted CT values of <20 (n = 14), <25 (n = 57), and <30 (n = 88), the RADT reached sensitivities of 100, 98.25, and 88.64%, respectively. All 29 culture-positive samples were detected by the RADT. Although the overall sensitivity was low, the Standard Q COVID-19 Ag test reliably detected patients with high RNA loads. In addition, negative RADT results fully corresponded with the lack of viral cultivability in Vero E6 cells. These results indicate that RADT can be a valuable tool for the detection of individuals with high RNA loads that are likely to transmit SARS-CoV-2.
Keywords: RT-qPCR; SARS-CoV-2; antigen test; infectiousness; virus culture.
Figures



References
-
- Hellewell J, Abbott S, Gimma A, Bosse NI, Jarvis CI, Russell TW, Munday JD, Kucharski AJ, Edmunds WJ, Funk S, Eggo RM, Sun F, Flasche S, Quilty BJ, Davies N, Liu Y, Clifford S, Klepac P, Jit M, Diamond C, Gibbs H, van Zandvoort K. 2020. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob Health 8:e488–e496. 10.1016/S2214-109X(20)30074-7. - DOI - PMC - PubMed
-
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 25:20000045. 10.2807/1560-7917.ES.2020.25.3.2000045. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

