Phosphorylation of the HP1β hinge region sequesters KAP1 in heterochromatin and promotes the exit from naïve pluripotency
- PMID: 34214177
- PMCID: PMC8287961
- DOI: 10.1093/nar/gkab548
Phosphorylation of the HP1β hinge region sequesters KAP1 in heterochromatin and promotes the exit from naïve pluripotency
Abstract
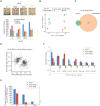
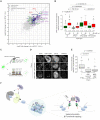
Heterochromatin binding protein HP1β plays an important role in chromatin organization and cell differentiation, however the underlying mechanisms remain unclear. Here, we generated HP1β-/- embryonic stem cells and observed reduced heterochromatin clustering and impaired differentiation. We found that during stem cell differentiation, HP1β is phosphorylated at serine 89 by CK2, which creates a binding site for the pluripotency regulator KAP1. This phosphorylation dependent sequestration of KAP1 in heterochromatin compartments causes a downregulation of pluripotency factors and triggers pluripotency exit. Accordingly, HP1β-/- and phospho-mutant cells exhibited impaired differentiation, while ubiquitination-deficient KAP1-/- cells had the opposite phenotype with enhanced differentiation. These results suggest that KAP1 regulates pluripotency via its ubiquitination activity. We propose that the formation of subnuclear membraneless heterochromatin compartments may serve as a dynamic reservoir to trap or release cellular factors. The sequestration of essential regulators defines a novel and active role of heterochromatin in gene regulation and represents a dynamic mode of remote control to regulate cellular processes like cell fate decisions.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Eissenberg J.C., Elgin S.C.. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 2000; 10:204–210. - PubMed
-
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T.. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001; 410:120–124. - PubMed
-
- Jacobs S.A., Khorasanizadeh S.. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002; 295:2080–2083. - PubMed
-
- Nakayama J., Rice J.C., Strahl B.D., Allis C.D., Grewal S.I.. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001; 292:110–113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

