The BARCODE1 Pilot: a feasibility study of using germline single nucleotide polymorphisms to target prostate cancer screening
- PMID: 34214236
- PMCID: PMC9292247
- DOI: 10.1111/bju.15535
The BARCODE1 Pilot: a feasibility study of using germline single nucleotide polymorphisms to target prostate cancer screening
Abstract
Objectives: To assess the feasibility and uptake of a community-based prostate cancer (PCa) screening programme selecting men according to their genetic risk of PCa. To assess the uptake of PCa screening investigations by men invited for screening. The uptake of the pilot study would guide the opening of the larger BARCODE1 study recruiting 5000 men.
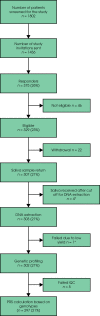
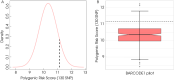
Subjects and methods: Healthy males aged 55-69 years were invited to participate via their general practitioners (GPs). Saliva samples were collected via mailed collection kits. After DNA extraction, genotyping was conducted using a study specific assay. Genetic risk was based on genotyping 130 germline PCa risk single nucleotide polymorphisms (SNPs). A polygenic risk score (PRS) was calculated for each participant using the sum of weighted alleles for 130 SNPs. Study participants with a PRS lying above the 90th centile value were invited for PCa screening by prostate magnetic resonance imaging (MRI) and biopsy.
Results: Invitation letters were sent to 1434 men. The overall study uptake was 26% (375/1436) and 87% of responders were eligible for study entry. DNA genotyping data were available for 297 men and 25 were invited for screening. After exclusions due to medical comorbidity/invitations declined, 18 of 25 men (72%) underwent MRI and biopsy of the prostate. There were seven diagnoses of PCa (38.9%). All cancers were low-risk and were managed with active surveillance.
Conclusion: The BARCODE1 Pilot has shown this community study in the UK to be feasible, with an overall uptake of 26%. The main BARCODE1 study is now open and will recruit 5000 men. The results of BARCODE1 will be important in defining the role of genetic profiling in targeted PCa population screening. Patient Summary What is the paper about? Very few prostate cancer screening programmes currently exist anywhere in the world. Our pilot study investigated if men in the UK would find it acceptable to have a genetic test based on a saliva sample to examine their risk of prostate cancer development. This test would guide whether men are offered prostate cancer screening tests. What does it mean for patients? We found that the study design was acceptable: 26% of men invited to take part agreed to have the test. The majority of men who were found to have an increased genetic risk of prostate cancer underwent further tests offered (prostate MRI scan and biopsy). We have now expanded the study to enrol 5000 men. The BARCODE1 study will be important in examining whether this approach could be used for large-scale population prostate cancer screening.
Keywords: GWAS; SNPs; precision medicine; prostate cancer; screening.
© 2021 The Authors BJU International published by John Wiley & Sons Ltd on behalf of BJU International.
Conflict of interest statement
Prof. Rosalind Eeles reports grants from European Research Council funding the submitted work. Prof Eeles reports personal fees from AstraZeneca UK Limited for her role as a member of an external Expert Committee as part of the Prostate Cancer Diagnosis Advisory Panel, and University of Chicago for an invited lecture, both of which are outside the submitted work. Dr Sarah Benafif, Dr Elizabeth Bancroft, Mrs Sarah Wakerell, Mrs Natalie Taylor, Mr Edward Saunders, Dr Holly Ni Raghallaigh, Ms Kathryn Myhill, Dr Eva McGrowder, Dr Zsofia Kote‐Jarai, Mr Denzil James, Mr Matthew Hogben, Mrs Lucia D’Mello, Mrs Barbara Benton, Dr Mark Brook, Mr Anthony Chamberlain, Mrs Reshma Rageevakuma and Dr Sibel Saya report funding for the submitted work from a European Research Council grant for this study. Dr Imran Rafi, Dr Tamsin Sevenoaks, Dr Andre Beattie, Dr Hywel Bowen‐Perkins, Dr Juliet Bower, Dr Shophia Kuganolipava report funding of research costs for the submitted work from The Institute of Cancer Research and from National Institute for Health Research: Clinical Research Network. Dr Michelle Ferris reports funding of research costs for the submitted work from The Institute of Cancer Research and from National Institute for Health Research: Clinical Research Network. Dr Ferris reports financial activities outside the submitted work related to the Befriend Your Boobs project for which she also has a patent, as well as a research grant from Noclor. Prof Antonis Antoniou is listed as creator of the BOADICEA algorithm that has been licensed by Cambridge Enterprise (outside the submitted work). The following authors have no conflict of interest to disclose: Dr Steve Hazell, Dr Christos Mikropoulos, Mr Pardeep Kumar, Prof Nandita De Souza.
Figures
Comment in
-
Prostate cancer screening: a new way forward or another false start?Nat Rev Urol. 2021 Oct;18(10):579-580. doi: 10.1038/s41585-021-00513-w. Nat Rev Urol. 2021. PMID: 34413502 No abstract available.
-
Polygenic risk scores and prostate cancer screening: a recipe for more overdiagnosis?BJU Int. 2022 Mar;129(3):271. doi: 10.1111/bju.15583. BJU Int. 2022. PMID: 35297162 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

