Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell
- PMID: 34214467
- PMCID: PMC8220945
- DOI: 10.1016/j.celrep.2021.109364
Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell
Abstract
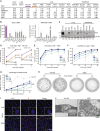
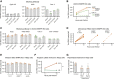
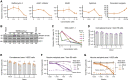
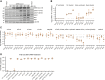
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) variants govern transmissibility, responsiveness to vaccination, and disease severity. In a screen for new models of SARS-CoV-2 infection, we identify human H522 lung adenocarcinoma cells as naturally permissive to SARS-CoV-2 infection despite complete absence of angiotensin-converting enzyme 2 (ACE2) expression. Remarkably, H522 infection requires the E484D S variant; viruses expressing wild-type S are not infectious. Anti-S monoclonal antibodies differentially neutralize SARS-CoV-2 E484D S in H522 cells as compared to ACE2-expressing cells. Sera from vaccinated individuals block this alternative entry mechanism, whereas convalescent sera are less effective. Although the H522 receptor remains unknown, depletion of surface heparan sulfates block H522 infection. Temporally resolved transcriptomic and proteomic profiling reveal alterations in cell cycle and the antiviral host cell response, including MDA5-dependent activation of type I interferon signaling. These findings establish an alternative SARS-CoV-2 host cell receptor for the E484D SARS-CoV-2 variant, which may impact tropism of SARS-CoV-2 and consequently human disease pathogenesis.
Keywords: ACE2-independent; COVID-19; RIG-I-like receptors; SARS-CoV-2; clathrin-mediated endocytosis; heparan sulfate; proteomics; spike variants; type I interferon; virus-host interactions.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.P.J.W., P.W.R., and Washington University have filed a patent application for uses of VSV-SARS-CoV-2. S.P.J.W. has received unrelated funding support in sponsored research agreements with Vir Biotechnology, Abbvie, and SAB therapeutics. M.S.D. is a consultant for Inbios, Vir Biotechnology, Fortress Biotech, and Carnival Corporation, and is on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology, Kaleido, and Emergent BioSolutions. J.E.C. has served as a consultant for Luna Biologics, is a member of the Scientific Advisory Boards of Meissa Vaccines, and is Founder of IDBiologics. The Crowe laboratory has received sponsored research agreements from Takeda, AstraZeneca, and IDBiologics. Vanderbilt University has applied for patents related to antibodies described in this paper. The remaining authors declare no competing interests.
Figures




Update of
-
Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell.bioRxiv [Preprint]. 2021 Mar 1:2021.03.01.433431. doi: 10.1101/2021.03.01.433431. bioRxiv. 2021. Update in: Cell Rep. 2021 Jul 13;36(2):109364. doi: 10.1016/j.celrep.2021.109364. PMID: 33688646 Free PMC article. Updated. Preprint.
References
-
- Agajanian M.J., Walker M.P., Axtman A.D., Ruela-de-Sousa R.R., Serafin D.S., Rabinowitz A.D., Graham D.M., Ryan M.B., Tamir T., Nakamichi Y. WNT activates the AAK1 kinase to promote clathrin-mediated endocytosis of LRP6 and establish a negative feedback loop. Cell Rep. 2019;26:79–93.e8. - PMC - PubMed
-
- Aguiar J.A., Tremblay B.J., Mansfield M.J., Woody O., Lobb B., Banerjee A., Chandiramohan A., Tiessen N., Cao Q., Dvorkin-Gheva A. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur. Respir. J. 2020;56:2001123. - PMC - PubMed
-
- Amraie R., Napoleon M.A., Yin W., Berrigan J., Suder E., Zhao G., Olejnik J., Gummuluru S., Muhlberger E., Chitalia V. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2 and are differentially expressed in lung and kidney epithelial and endothelial cells. bioRxiv. 2020 doi: 10.1101/2020.06.22.165803. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

