Genetic Determinants of Somatic Selection of Mutational Processes in 3,566 Human Cancers
- PMID: 34215622
- PMCID: PMC9662923
- DOI: 10.1158/0008-5472.CAN-21-0086
Genetic Determinants of Somatic Selection of Mutational Processes in 3,566 Human Cancers
Abstract
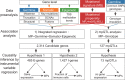
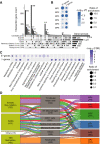
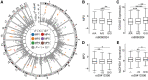
The somatic landscape of the cancer genome results from different mutational processes represented by distinct "mutational signatures." Although several mutagenic mechanisms are known to cause specific mutational signatures in cell lines, the variation of somatic mutational activities in patients, which is mostly attributed to somatic selection, is still poorly explained. Here, we introduce a quantitative trait, mutational propensity (MP), and describe an integrated method to infer genetic determinants of variations in the mutational processes in 3,566 cancers with specific underlying mechanisms. As a result, we report 2,314 candidate determinants with both significant germline and somatic effects on somatic selection of mutational processes, of which, 485 act via cancer gene expression and 1,427 act through the tumor-immune microenvironment. These data demonstrate that the genetic determinants of MPs provide complementary information to known cancer driver genes, clonal evolution, and clinical biomarkers. SIGNIFICANCE: The genetic determinants of the somatic mutational processes in cancer elucidate the biology underlying somatic selection and evolution of cancers and demonstrate complementary predictive power across cancer types.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

