Atomistic molecular dynamics simulations of tubulin heterodimers explain the motion of a microtubule
- PMID: 34215900
- PMCID: PMC8448678
- DOI: 10.1007/s00249-021-01553-1
Atomistic molecular dynamics simulations of tubulin heterodimers explain the motion of a microtubule
Abstract
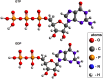
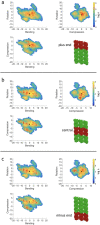
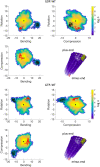
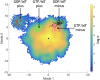
Microtubules are essential parts of the cytoskeleton that are built by polymerization of tubulin heterodimers into a hollow tube. Regardless that their structures and functions have been comprehensively investigated in a modern soft matter, it is unclear how properties of tubulin heterodimer influence and promote the self-assembly. A detailed knowledge of such structural mechanisms would be helpful in drug design against neurodegenerative diseases, cancer, diabetes etc. In this work atomistic molecular dynamics simulations were used to investigate the fundamental dynamics of tubulin heterodimers in a sheet and a short microtubule utilizing well-equilibrated structures. The breathing motions of the tubulin heterodimers during assembly show that the movement at the lateral interface between heterodimers (wobbling) dominates in the lattice. The simulations of the protofilament curvature agrees well with recently published experimental data, showing curved protofilaments at polymerization of the microtubule plus end. The tubulin heterodimers exposed at the microtubule minus end were less curved and displayed altered interactions at the site of sheet closure around the outmost heterodimers, which may slow heterodimer binding and polymerization, providing a potential explanation for the limited dynamics observed at the minus end.
Keywords: Alzheimer’s disease; Diabetes; Equilibrium dynamics; Microtubules; Molecular dynamics; Protofilament; Small drugs.
© 2021. The Author(s).
Conflict of interest statement
There are no conflicts to declare.
Figures


Similar articles
-
Cypin binds to tubulin heterodimers and microtubule protofilaments and regulates microtubule spacing in developing hippocampal neurons.Mol Cell Neurosci. 2022 Dec;123:103783. doi: 10.1016/j.mcn.2022.103783. Epub 2022 Oct 5. Mol Cell Neurosci. 2022. PMID: 36208859 Free PMC article.
-
Intrinsic bending of microtubule protofilaments.Structure. 2011 Mar 9;19(3):409-17. doi: 10.1016/j.str.2010.12.020. Structure. 2011. PMID: 21397191
-
Mutations in Human Tubulin Proximal to the Kinesin-Binding Site Alter Dynamic Instability at Microtubule Plus- and Minus-Ends.Dev Cell. 2016 Apr 4;37(1):72-84. doi: 10.1016/j.devcel.2016.03.003. Dev Cell. 2016. PMID: 27046833 Free PMC article.
-
How tubulin subunits are lost from the shortening ends of microtubules.J Struct Biol. 1997 Mar;118(2):107-18. doi: 10.1006/jsbi.1997.3844. J Struct Biol. 1997. PMID: 9126637 Review.
-
Tipping microtubule dynamics, one protofilament at a time.Curr Opin Cell Biol. 2018 Feb;50:86-93. doi: 10.1016/j.ceb.2018.02.015. Epub 2018 Mar 21. Curr Opin Cell Biol. 2018. PMID: 29573640 Review.
References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

