Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency
- PMID: 34216544
- PMCID: PMC9089372
- DOI: 10.1016/j.molcel.2021.06.011
Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency
Erratum in
-
Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency.Mol Cell. 2021 Aug 5;81(15):3227. doi: 10.1016/j.molcel.2021.07.015. Mol Cell. 2021. PMID: 34358459 No abstract available.
Abstract
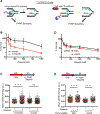
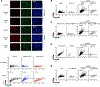
Mutations in BRCA1 or BRCA2 (BRCA) is synthetic lethal with poly(ADP-ribose) polymerase inhibitors (PARPi). Lethality is thought to derive from DNA double-stranded breaks (DSBs) necessitating BRCA function in homologous recombination (HR) and/or fork protection (FP). Here, we report instead that toxicity derives from replication gaps. BRCA1- or FANCJ-deficient cells, with common repair defects but distinct PARPi responses, reveal gaps as a distinguishing factor. We further uncouple HR, FP, and fork speed from PARPi response. Instead, gaps characterize BRCA-deficient cells, are diminished upon resistance, restored upon resensitization, and, when exposed, augment PARPi toxicity. Unchallenged BRCA1-deficient cells have elevated poly(ADP-ribose) and chromatin-associated PARP1, but aberrantly low XRCC1 consistent with defects in backup Okazaki fragment processing (OFP). 53BP1 loss resuscitates OFP by restoring XRCC1-LIG3 that suppresses the sensitivity of BRCA1-deficient cells to drugs targeting OFP or generating gaps. We highlight gaps as a determinant of PARPi toxicity changing the paradigm for synthetic lethal interactions.
Keywords: BRCA1/BRCA2; Fanconi anemia (FA); Okazaki fragment processing; PARP inhibitor; fork protection; homologous recombination; parylation; replication gaps; ssDNA; synthetic lethal.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Amé JC, Fouquerel E, Gauthier LR, Biard D, Boussin FD, Dantzer F, de Murcia G, and Schreiber V (2009). Radiation-induced mitotic catastrophe in PARG-deficient cells. J. Cell Sci 122, 1990–2002. - PubMed
-
- Avkin S, Sevilya Z, Toube L, Geacintov N, Chaney SG, Oren M, and Livneh Z (2006). p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell 22, 407–413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

