Machine learning reveals sequence-function relationships in family 7 glycoside hydrolases
- PMID: 34216620
- PMCID: PMC8329511
- DOI: 10.1016/j.jbc.2021.100931
Machine learning reveals sequence-function relationships in family 7 glycoside hydrolases
Abstract
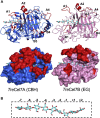
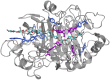
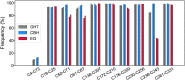
Family 7 glycoside hydrolases (GH7) are among the principal enzymes for cellulose degradation in nature and industrially. These enzymes are often bimodular, including a catalytic domain and carbohydrate-binding module (CBM) attached via a flexible linker, and exhibit an active site that binds cello-oligomers of up to ten glucosyl moieties. GH7 cellulases consist of two major subtypes: cellobiohydrolases (CBH) and endoglucanases (EG). Despite the critical importance of GH7 enzymes, there remain gaps in our understanding of how GH7 sequence and structure relate to function. Here, we employed machine learning to gain data-driven insights into relationships between sequence, structure, and function across the GH7 family. Machine-learning models, trained only on the number of residues in the active-site loops as features, were able to discriminate GH7 CBHs and EGs with up to 99% accuracy, demonstrating that the lengths of loops A4, B2, B3, and B4 strongly correlate with functional subtype across the GH7 family. Classification rules were derived such that specific residues at 42 different sequence positions each predicted the functional subtype with accuracies surpassing 87%. A random forest model trained on residues at 19 positions in the catalytic domain predicted the presence of a CBM with 89.5% accuracy. Our machine learning results recapitulate, as top-performing features, a substantial number of the sequence positions determined by previous experimental studies to play vital roles in GH7 activity. We surmise that the yet-to-be-explored sequence positions among the top-performing features also contribute to GH7 functional variation and may be exploited to understand and manipulate function.
Keywords: Trichoderma reesei; bioinformatics; cellulase; glycoside hydrolase; statistics; tryptophan.
Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures







References
-
- Himmel M.E., Ding S.Y., Johnson D.K., Adney W.S., Nimlos M.R., Brady J.W., Foust T.D. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. - PubMed
-
- Payne C.M., Knott B.C., Mayes H.B., Hansson H., Himmel M.E., Sandgren M., Stahlberg J., Beckham G.T. Fungal cellulases. Chem. Rev. 2015;115:1308–1448. - PubMed
-
- Zhang Y.H.P., Lynd L.R. Toward an aggregated understanding of enzymatic hydrolysis of cellulose: Noncomplexed cellulase systems. Biotech. Bioeng. 2004;88:797–824. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

