A systematic evaluation of source reconstruction of resting MEG of the human brain with a new high-resolution atlas: Performance, precision, and parcellation
- PMID: 34219311
- PMCID: PMC8410546
- DOI: 10.1002/hbm.25578
A systematic evaluation of source reconstruction of resting MEG of the human brain with a new high-resolution atlas: Performance, precision, and parcellation
Abstract
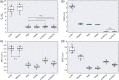
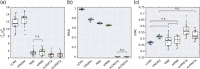
Noninvasive functional neuroimaging of the human brain can give crucial insight into the mechanisms that underpin healthy cognition and neurological disorders. Magnetoencephalography (MEG) measures extracranial magnetic fields originating from neuronal activity with high temporal resolution, but requires source reconstruction to make neuroanatomical inferences from these signals. Many source reconstruction algorithms are available, and have been widely evaluated in the context of localizing task-evoked activities. However, no consensus yet exists on the optimum algorithm for resting-state data. Here, we evaluated the performance of six commonly-used source reconstruction algorithms based on minimum-norm and beamforming estimates. Using human resting-state MEG, we compared the algorithms using quantitative metrics, including resolution properties of inverse solutions and explained variance in sensor-level data. Next, we proposed a data-driven approach to reduce the atlas from the Human Connectome Project's multi-modal parcellation of the human cortex based on metrics such as MEG signal-to-noise-ratio and resting-state functional connectivity gradients. This procedure produced a reduced cortical atlas with 230 regions, optimized to match the spatial resolution and the rank of MEG data from the current generation of MEG scanners. Our results show that there is no "one size fits all" algorithm, and make recommendations on the appropriate algorithms depending on the data and aimed analyses. Our comprehensive comparisons and recommendations can serve as a guide for choosing appropriate methodologies in future studies of resting-state MEG.
Keywords: Magnetoencephalography; cortical atlas parcellation; resolution analysis; resting-state; source reconstruction.
© 2021 The Authors. Human Brain Mapping published by Wiley Periodicals LLC.
Figures





Similar articles
-
A human brain atlas derived via n-cut parcellation of resting-state and task-based fMRI data.Magn Reson Imaging. 2016 Feb;34(2):209-18. doi: 10.1016/j.mri.2015.10.036. Epub 2015 Oct 31. Magn Reson Imaging. 2016. PMID: 26523655 Free PMC article.
-
Adaptive cortical parcellations for source reconstructed EEG/MEG connectomes.Neuroimage. 2018 Apr 1;169:23-45. doi: 10.1016/j.neuroimage.2017.09.009. Epub 2017 Sep 8. Neuroimage. 2018. PMID: 28893608 Free PMC article.
-
Reproducibility of graph measures derived from resting-state MEG functional connectivity metrics in sensor and source spaces.Hum Brain Mapp. 2022 Mar;43(4):1342-1357. doi: 10.1002/hbm.25726. Epub 2022 Jan 12. Hum Brain Mapp. 2022. PMID: 35019189 Free PMC article.
-
Magnetoencephalography Signal Processing, Forward Modeling, Magnetoencephalography Inverse Source Imaging, and Coherence Analysis.Neuroimaging Clin N Am. 2020 May;30(2):125-143. doi: 10.1016/j.nic.2020.02.001. Neuroimaging Clin N Am. 2020. PMID: 32336402 Review.
-
Magnetoencephalography for localizing and characterizing the epileptic focus.Handb Clin Neurol. 2019;160:203-214. doi: 10.1016/B978-0-444-64032-1.00013-8. Handb Clin Neurol. 2019. PMID: 31277848 Review.
Cited by
-
Dementia ConnEEGtome: Towards multicentric harmonization of EEG connectivity in neurodegeneration.Int J Psychophysiol. 2022 Feb;172:24-38. doi: 10.1016/j.ijpsycho.2021.12.008. Epub 2021 Dec 27. Int J Psychophysiol. 2022. PMID: 34968581 Free PMC article. Review.
-
Oscillatory characteristics of resting-state magnetoencephalography reflect pathological and symptomatic conditions of cognitive impairment.Front Aging Neurosci. 2024 Jan 30;16:1273738. doi: 10.3389/fnagi.2024.1273738. eCollection 2024. Front Aging Neurosci. 2024. PMID: 38352236 Free PMC article.
-
Dorsal brain activity reflects the severity of menopausal symptoms.Menopause. 2024 May 1;31(5):399-407. doi: 10.1097/GME.0000000000002347. Epub 2024 Apr 15. Menopause. 2024. PMID: 38626372 Free PMC article.
-
MEG cortical microstates: Spatiotemporal characteristics, dynamic functional connectivity and stimulus-evoked responses.Neuroimage. 2022 May 1;251:119006. doi: 10.1016/j.neuroimage.2022.119006. Epub 2022 Feb 16. Neuroimage. 2022. PMID: 35181551 Free PMC article.
-
Whole-Brain Network Models: From Physics to Bedside.Front Comput Neurosci. 2022 May 26;16:866517. doi: 10.3389/fncom.2022.866517. eCollection 2022. Front Comput Neurosci. 2022. PMID: 35694610 Free PMC article. Review.
References
-
- Babiloni, C., Lizio, R., Marzano, N., Capotosto, P., Soricelli, A., Triggiani, A., … Del Percio, C. (2016). Brain neural synchronization and functional coupling in Alzheimer's disease as revealed by resting state EEG rhythms. International Journal of Psychophysiology, 103, 88–102. 10.1016/j.ijpsycho.2015.02.008 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

