Structure and Function of Major SARS-CoV-2 and SARS-CoV Proteins
- PMID: 34220199
- PMCID: PMC8221690
- DOI: 10.1177/11779322211025876
Structure and Function of Major SARS-CoV-2 and SARS-CoV Proteins
Abstract
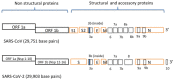
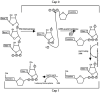
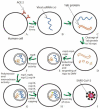
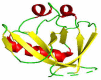
SARS-CoV-2 virus, the causative agent of COVID-19 pandemic, has a genomic organization consisting of 16 nonstructural proteins (nsps), 4 structural proteins, and 9 accessory proteins. Relative of SARS-CoV-2, SARS-CoV, has genomic organization, which is very similar. In this article, the function and structure of the proteins of SARS-CoV-2 and SARS-CoV are described in great detail. The nsps are expressed as a single or two polyproteins, which are then cleaved into individual proteins using two proteases of the virus, a chymotrypsin-like protease and a papain-like protease. The released proteins serve as centers of virus replication and transcription. Some of these nsps modulate the host's translation and immune systems, while others help the virus evade the host immune system. Some of the nsps help form replication-transcription complex at double-membrane vesicles. Others, including one RNA-dependent RNA polymerase and one exonuclease, help in the polymerization of newly synthesized RNA of the virus and help minimize the mutation rate by proofreading. After synthesis of the viral RNA, it gets capped. The capping consists of adding GMP and a methylation mark, called cap 0 and additionally adding a methyl group to the terminal ribose called cap1. Capping is accomplished with the help of a helicase, which also helps remove a phosphate, two methyltransferases, and a scaffolding factor. Among the structural proteins, S protein forms the receptor of the virus, which latches on the angiotensin-converting enzyme 2 receptor of the host and N protein binds and protects the genomic RNA of the virus. The accessory proteins found in these viruses are small proteins with immune modulatory roles. Besides functions of these proteins, solved X-ray and cryogenic electron microscopy structures related to the function of the proteins along with comparisons to other coronavirus homologs have been described in the article. Finally, the rate of mutation of SARS-CoV-2 residues of the proteome during the 2020 pandemic has been described. Some proteins are mutated more often than other proteins, but the significance of these mutation rates is not fully understood.
Keywords: SARS-CoV; SARS-CoV-2; function; proteins; structure.
© The Author(s) 2021.
Conflict of interest statement
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures





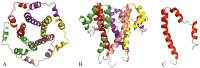
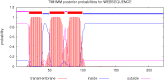


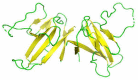
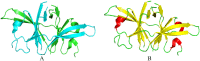
Similar articles
-
Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.Mol Biomed. 2020;1(1):5. doi: 10.1186/s43556-020-00005-0. Epub 2020 Aug 20. Mol Biomed. 2020. PMID: 34765992 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
Potential 3-chymotrypsin-like cysteine protease cleavage sites in the coronavirus polyproteins pp1a and pp1ab and their possible relevance to COVID-19 vaccine and drug development.FASEB J. 2021 May;35(5):e21573. doi: 10.1096/fj.202100280RR. FASEB J. 2021. PMID: 33913206 Free PMC article. Review.
-
1H, 13C, and 15N backbone chemical shift assignments of the nucleic acid-binding domain of SARS-CoV-2 non-structural protein 3e.Biomol NMR Assign. 2020 Oct;14(2):329-333. doi: 10.1007/s12104-020-09971-6. Epub 2020 Aug 8. Biomol NMR Assign. 2020. PMID: 32770392 Free PMC article.
-
Structure of Nonstructural Protein 1 from SARS-CoV-2.J Virol. 2021 Jan 28;95(4):e02019-20. doi: 10.1128/JVI.02019-20. Print 2021 Jan 28. J Virol. 2021. PMID: 33234675 Free PMC article.
Cited by
-
Genetic diversity and evolution of porcine hemagglutinating encephalomyelitis virus in Guangxi province of China during 2021-2024.Front Microbiol. 2024 Oct 9;15:1474552. doi: 10.3389/fmicb.2024.1474552. eCollection 2024. Front Microbiol. 2024. PMID: 39444682 Free PMC article.
-
Predicting the spread of SARS-CoV-2 variants: An artificial intelligence enabled early detection.PNAS Nexus. 2024 Jan 2;3(1):pgad424. doi: 10.1093/pnasnexus/pgad424. eCollection 2024 Jan. PNAS Nexus. 2024. PMID: 38170049 Free PMC article.
-
Modelling the Transitioning of SARS-CoV-2 nsp3 and nsp4 Lumenal Regions towards a More Stable State on Complex Formation.Int J Mol Sci. 2022 Dec 31;24(1):720. doi: 10.3390/ijms24010720. Int J Mol Sci. 2022. PMID: 36614163 Free PMC article.
-
Adenovirus 5 Vectors Expressing SARS-CoV-2 Proteins.mSphere. 2022 Apr 27;7(2):e0099821. doi: 10.1128/msphere.00998-21. Epub 2022 Feb 28. mSphere. 2022. PMID: 35224978 Free PMC article.
-
Theophylline: Old Drug in a New Light, Application in COVID-19 through Computational Studies.Int J Mol Sci. 2022 Apr 9;23(8):4167. doi: 10.3390/ijms23084167. Int J Mol Sci. 2022. PMID: 35456985 Free PMC article. Review.
References
-
- Marra MA, Jones SJM, Astell CR, et al.. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399-1404. doi:10.1126/science.1085953. - PubMed
-
- Rota PA, Oberste MS, Monroe SS, et al.. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394-1399. doi:10.1126/science.1085952. - PubMed
-
- Thiel V, Ivanov KA, Putics Á, et al.. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305-2315. doi:10.1099/vir.0.19424-0. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

