Use of plant growth-promoting bacteria to enhance salinity stress in soybean (Glycine max L.) plants
- PMID: 34220237
- PMCID: PMC8241701
- DOI: 10.1016/j.sjbs.2021.03.053
Use of plant growth-promoting bacteria to enhance salinity stress in soybean (Glycine max L.) plants
Abstract
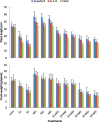
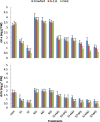
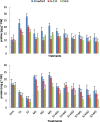
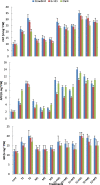
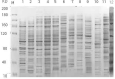
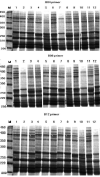
The effects of three rhizobacterial isolates namely Pseudomonas fluorescens (M1), Pseudomonas putida (M2) and Bacillus subtilis (M3) were examined to enhance growth and chemical components such as chlorophyll and proline of three cultivars of soybean (Glycine max L.) under two levels of salinity stress (S1 = 200 mM and S2 = 400 mM of NaCl salt). Several morphological and physiological parameters were investigated. The highest mean values of final germination percent (FGP) were registered in cultivar Crawford (95%) followed by Giza111 cultivar (93%) in the presence of P. fluorescens, while, FGP of Clark was 85%. Mean germination time was decreased by the application of P. fluorescens or P. putida in both salt stressed and unstressed traits. All growth parameters were significantly decreased by salinity treatments, particularly at S2. A significant increase in stem length and shoot fresh weight was recorded in plants treated with P. fluorescens. This enhancing trend was followed by the application of P. putida then B. subtilis. Chlorophyll contents and plant soluble proteins were decreased, while proline content was increased as compared with control treatment. Results showed that the salt tolerant cultivar, Crawford, may have a better tolerance strategy against oxidative damages by increasing antioxidant enzymes activities under high salinity stress. These results suggest that salt induced oxidative stress in soybean is generally counteracted by enzymatic defense systems stimulated under harsh conditions. Our results showed that inoculation with plant growth-promoting rhizobacterial (PGPR) alleviated the harmful effects of salinity stress on soybean cultivars. The diversity in the phylogenetic relationship and in the level of genetic among cultivars was assessed by SDS-PAGE and RAPD markers. Among the polymorphism bands, only few were found to be useful as positive or negative markers associated with salt stress. The maximum number of bands (17) was recorded in Crawford, while the minimum number of bands (11) was recorded in Clark. Therefore, the ISSR can be used to identify alleles associated with the salt stress in soybean germplasm.
Keywords: Antioxidant enzymes; Plant growth promoting rhizobacteria (PGPR); Protein banding; Salt stress; Soybean.
© 2021 The Authors.
Conflict of interest statement
The authors declare that there is no conflict of interests regarding the publications of this paper.
Figures
References
-
- Abdelmongy, A.S., El-Moselhy, K. 2015. Seasonal variation of the physical and chemical properties of seawater at the Northern Red Sea, Egypt. Open J. Ocean and Coastal Scie. 2(1), ISSN:2377-0007. DOI:10.15764/OCS.2015.2015.01001.
-
- Akyol, T., Yılmaz, O., Uzilday, B., Ozgur, Turkan, I. 2020. Plant response to salinity: An analysis of ROS formation, signaling, and antioxidant defense. Turkish J. Bot. 44: 1-13. doi10.3906/bot-1911-15.
-
- Albacete, A., Ghanem, M.E., Martínez-Andújar, C., Acosta, M., Sánchez-Bravo, J., Martínez, V., Stanley, L., Ian, C. Dodd, P.F. 2008. Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Botany. 59(15): 4119-4131. - PMC - PubMed
-
- Bates, L.S., Waldren, R.P., Teare, I.D. 1973. Rapid determination of free proline for water-stress studies. Plant Soil. 39: 205–207.
-
- Benidire L., Oufdou K., Barakat M., Ortet P., Heulin T., Achouak W. Phytobeneficial bacteria improve saline stress tolerance in Vicia faba and modulate microbial interaction network. Sci. Tot. Environ. 2020;729 - PubMed
LinkOut - more resources
Full Text Sources

