An Integrated Database of Small RNAs and Their Interplay With Transcriptional Gene Regulatory Networks in Corynebacteria
- PMID: 34220744
- PMCID: PMC8247434
- DOI: 10.3389/fmicb.2021.656435
An Integrated Database of Small RNAs and Their Interplay With Transcriptional Gene Regulatory Networks in Corynebacteria
Abstract
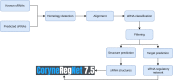
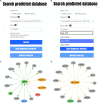
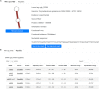
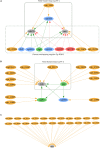
Small RNAs (sRNAs) are one of the key players in the post-transcriptional regulation of bacterial gene expression. These molecules, together with transcription factors, form regulatory networks and greatly influence the bacterial regulatory landscape. Little is known concerning sRNAs and their influence on the regulatory machinery in the genus Corynebacterium, despite its medical, veterinary and biotechnological importance. Here, we expand corynebacterial regulatory knowledge by integrating sRNAs and their regulatory interactions into the transcriptional regulatory networks of six corynebacterial species, covering four human and animal pathogens, and integrate this data into the CoryneRegNet database. To this end, we predicted sRNAs to regulate 754 genes, including 206 transcription factors, in corynebacterial gene regulatory networks. Amongst them, the sRNA Cd-NCTC13129-sRNA-2 is predicted to directly regulate ydfH, which indirectly regulates 66 genes, including the global regulator glxR in C. diphtheriae. All of the sRNA-enriched regulatory networks of the genus Corynebacterium have been made publicly available in the newest release of CoryneRegNet(www.exbio.wzw.tum.de/coryneregnet/) to aid in providing valuable insights and to guide future experiments.
Keywords: CoryneRegNet; Corynebacterium; gene regulatory networks; sRNA targets; small RNAs.
Copyright © 2021 Parise, Parise, Aburjaile, Pinto Gomide, Kato, Raden, Backofen, Azevedo and Baumbach.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
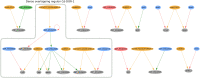
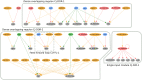
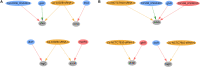
References
-
- Ahmed W., Hafeez M. A., Mahmood S. (2018). Identification and functional characterization of bacterial small non-coding RNAs and their target: a review. Gene Rep. 10 167–176. 10.1016/j.genrep.2018.01.001 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

