Molecular Characterization and Survive Abilities of Salmonella Heidelberg Strains of Poultry Origin in Brazil
- PMID: 34220757
- PMCID: PMC8253257
- DOI: 10.3389/fmicb.2021.674147
Molecular Characterization and Survive Abilities of Salmonella Heidelberg Strains of Poultry Origin in Brazil
Abstract
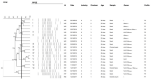
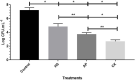
The aim of the study was to evaluate the genotypic and phenotypic characteristics of 20 strains of S. Heidelberg (SH) isolated from broilers produced in southern Brazil. The similarity and presence of genetic determinants linked to virulence, antimicrobial resistance, biofilm formation, and in silico-predicted metabolic interactions revealed this serovar as a threat to public health. The presence of the ompC, invA, sodC, avrA, lpfA, and agfA genes was detected in 100% of the strains and the luxS gene in 70% of them. None of the strains carries the bla SHV, mcr-1, qnrA, qnrB, and qnrS genes. All strains showed a multidrug-resistant profile to at least three non-β-lactam drugs, which include colistin, sulfamethoxazole, and tetracycline. Resistance to penicillin, ceftriaxone (90%), meropenem (25%), and cefoxitin (25%) were associated with the presence of bla CTX-M and bla CMY-2 genes. Biofilm formation reached a mature stage at 25 and 37°C, especially with chicken juice (CJ) addition. The sodium hypochlorite 1% was the least efficient in controlling the sessile cells. Genomic analysis of two strains identified more than 100 virulence genes and the presence of resistance to 24 classes of antibiotics correlated to phenotypic tests. Protein-protein interaction (PPI) prediction shows two metabolic pathways correlation with biofilm formation. Virulence, resistance, and biofilm determinants must be constant monitoring in SH, due to the possibility of occurring infections extremely difficult to cure and due risk of the maintenance of the bacterium in production environments.
Keywords: antimicrobial resistance; biofilms; salmonellosis; virulence; whole genome sequencing.
Copyright © 2021 Melo, Galvão, Guidotti-Takeuchi, Peres, Fonseca, Profeta, Azevedo, Monteiro, Brenig and Rossi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Phenotypic and Genotypic Characterization of Virulence Factors and Susceptibility to Antibiotics in Salmonella Infantis Strains Isolated from Chicken Meat: First Findings in Chile.Animals (Basel). 2020 Jun 18;10(6):1049. doi: 10.3390/ani10061049. Animals (Basel). 2020. PMID: 32570768 Free PMC article.
-
[Characterization of Salmonella Heidelberg strains isolated in Chile].Rev Med Chil. 2019;147(1):24-33. doi: 10.4067/S0034-98872019000100024. Rev Med Chil. 2019. PMID: 30848761 Spanish.
-
Phenotypic and Genomic Assessment of Antimicrobial Resistance and Virulence Factors Determinants in Salmonella Heidelberg Isolated from Broiler Chickens.Animals (Basel). 2025 Mar 30;15(7):1003. doi: 10.3390/ani15071003. Animals (Basel). 2025. PMID: 40218396 Free PMC article.
-
A Whole-Genome Sequencing Approach To Study Cefoxitin-Resistant Salmonella enterica Serovar Heidelberg Isolates from Various Sources.Antimicrob Agents Chemother. 2017 Mar 24;61(4):e01919-16. doi: 10.1128/AAC.01919-16. Print 2017 Apr. Antimicrob Agents Chemother. 2017. PMID: 28137797 Free PMC article.
-
Phenotypic and Molecular Characterization of Salmonella Enteritidis SE86 Isolated from Poultry and Salmonellosis Outbreaks.Foodborne Pathog Dis. 2017 Dec;14(12):742-754. doi: 10.1089/fpd.2017.2327. Epub 2017 Nov 6. Foodborne Pathog Dis. 2017. PMID: 29106298
Cited by
-
Antimicrobial resistance and genetic background of non-typhoidal Salmonella enterica strains isolated from human infections in São Paulo, Brazil (2000-2019).Braz J Microbiol. 2022 Sep;53(3):1249-1262. doi: 10.1007/s42770-022-00748-8. Epub 2022 Apr 21. Braz J Microbiol. 2022. PMID: 35446010 Free PMC article.
-
Effect of diets containing commercial bioactive compounds on Salmonella Heidelberg infection in broiler chicks.Braz J Microbiol. 2023 Mar;54(1):571-577. doi: 10.1007/s42770-022-00899-8. Epub 2022 Dec 27. Braz J Microbiol. 2023. PMID: 36572822 Free PMC article.
-
Global trends in typhoid and paratyphoid, and invasive non-typhoidal salmonella, and the burden of antimicrobial resistance: a trend analysis study from 1990 to 2021.Front Med (Lausanne). 2025 May 20;12:1588507. doi: 10.3389/fmed.2025.1588507. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40463975 Free PMC article.
-
A Ternary Copper (II) Complex with 4-Fluorophenoxyacetic Acid Hydrazide in Combination with Antibiotics Exhibits Positive Synergistic Effect against Salmonella Typhimurium.Antibiotics (Basel). 2022 Mar 15;11(3):388. doi: 10.3390/antibiotics11030388. Antibiotics (Basel). 2022. PMID: 35326852 Free PMC article.
-
Interference with Bacterial Conjugation and Natural Alternatives to Antibiotics: Bridging a Gap.Antibiotics (Basel). 2023 Jun 29;12(7):1127. doi: 10.3390/antibiotics12071127. Antibiotics (Basel). 2023. PMID: 37508224 Free PMC article.
References
-
- ABPA (2020). Relatório Anual Assoc. Bras. Prote na Anim. 160. Available online at: http://abpa-br.org/relatorios/
-
- Alcántar-Curiel M. D., Ledezma-Escalante C. A., Jarillo-Quijada M. D., Gayosso-Vázquez C., Morfín-Otero R., Rodríguez-Noriega E., et al. (2018). Association of antibiotic resistance, cell adherence, and biofilm production with the endemicity of nosocomial Klebsiella pneumoniae. Biomed Res. Int. 2018 1–9. 10.1155/2018/7012958 - DOI - PMC - PubMed
-
- Allan Pfuntner (2011). Sanitizers and disinfectants: The chemicals of prevention. Food Saf. Mag. Available online at: https://www.food-safety.com/articles/6707-sanitizers-and-disinfectants-t....
-
- Alzwghaibi A. B., Yahyaraeyat R., Fasaei B. N., Langeroudi A. G., Salehi T. Z. (2018). Rapid molecular identification and differentiation of common Salmonella serovars isolated from poultry, domestic animals and foodstuff using multiplex PCR assay. Arch. Microbiol. 200 1009–1016. 10.1007/s00203-018-1501-1507 - DOI - PubMed
-
- Amini K., Salehi T. Z., Nikbakht G., Ranjbar R., Amini J. (2010). Molecular detection of invA and spv virulence genes in. Salmonella enteritidis isolated from human and animals in Iran. African J. Microbiol. Res. 4 2202–2210.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

