First Report of Coexistence of bla SFO-1 and bla NDM-1 β-Lactamase Genes as Well as Colistin Resistance Gene mcr-9 in a Transferrable Plasmid of a Clinical Isolate of Enterobacter hormaechei
- PMID: 34220761
- PMCID: PMC8252965
- DOI: 10.3389/fmicb.2021.676113
First Report of Coexistence of bla SFO-1 and bla NDM-1 β-Lactamase Genes as Well as Colistin Resistance Gene mcr-9 in a Transferrable Plasmid of a Clinical Isolate of Enterobacter hormaechei
Erratum in
-
Corrigendum: First Report of Coexistence of bla SFO-1 and bla NDM-1 β-Lactamase Genes as Well as Colistin Resistance Gene mcr-9 in a Transferrable Plasmid of a Clinical Isolate of Enterobacter hormaechei.Front Microbiol. 2021 Sep 28;12:741628. doi: 10.3389/fmicb.2021.741628. eCollection 2021. Front Microbiol. 2021. PMID: 34650541 Free PMC article.
Abstract
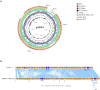
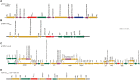
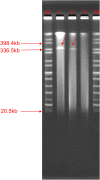
Many antimicrobial resistance genes usually located on transferable plasmids are responsible for multiple antimicrobial resistance among multidrug-resistant (MDR) Gram-negative bacteria. The aim of this study is to characterize a carbapenemase-producing Enterobacter hormaechei 1575 isolate from the blood sample in a tertiary hospital in Wuhan, Hubei Province, China. Antimicrobial susceptibility test showed that 1575 was an MDR isolate. The whole genome sequencing (WGS) and comparative genomics were used to deeply analyze the molecular information of the 1575 and to explore the location and structure of antibiotic resistance genes. The three key resistance genes (bla SFO-1, bla NDM-1, and mcr-9) were verified by PCR, and the amplicons were subsequently sequenced. Moreover, the conjugation assay was also performed to determine the transferability of those resistance genes. Plasmid files were determined by the S1 nuclease pulsed-field gel electrophoresis (S1-PFGE). WGS revealed that p1575-1 plasmid was a conjugative plasmid that possessed the rare coexistence of bla SFO-1, bla NDM-1, and mcr-9 genes and complete conjugative systems. And p1575-1 belonged to the plasmid incompatibility group IncHI2 and multilocus sequence typing ST102. Meanwhile, the pMLST type of p1575-1 was IncHI2-ST1. Conjugation assay proved that the MDR p1575-1 plasmid could be transferred to other recipients. S1-PFGE confirmed the location of plasmid with molecular weight of 342,447 bp. All these three resistant genes were flanked by various mobile elements, indicating that the bla SFO-1, bla NDM-1, and mcr-9 could be transferred not only by the p1575-1 plasmid but also by these mobile elements. Taken together, we report for the first time the coexistence of bla SFO-1, bla NDM-1, and mcr-9 on a transferable plasmid in a MDR clinical isolate E. hormaechei, which indicates the possibility of horizontal transfer of antibiotic resistance genes.
Keywords: Enterobacter hormaechei; IncHI2; WGS; blaNDM–1; blaSFO–1; mcr-9; mobile elements; plasmid.
Copyright © 2021 Ai, Zhou, Wang, Zhan, Hu, Xu, Guo, Wang, Yu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
First report of coexistence of bla KPC-2-, bla NDM-1- and mcr-9-carrying plasmids in a clinical carbapenem-resistant Enterobacter hormaechei isolate.Front Microbiol. 2023 Mar 23;14:1153366. doi: 10.3389/fmicb.2023.1153366. eCollection 2023. Front Microbiol. 2023. PMID: 37032905 Free PMC article.
-
Corrigendum: First Report of Coexistence of bla SFO-1 and bla NDM-1 β-Lactamase Genes as Well as Colistin Resistance Gene mcr-9 in a Transferrable Plasmid of a Clinical Isolate of Enterobacter hormaechei.Front Microbiol. 2021 Sep 28;12:741628. doi: 10.3389/fmicb.2021.741628. eCollection 2021. Front Microbiol. 2021. PMID: 34650541 Free PMC article.
-
Molecular characterization of NDM-1-producing carbapenem-resistant E. cloacae complex from a tertiary hospital in Chongqing, China.Front Cell Infect Microbiol. 2022 Aug 8;12:935165. doi: 10.3389/fcimb.2022.935165. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36004335 Free PMC article.
-
Co-infections of two carbapenemase-producing Enterobacter hormaechei clinical strains isolated from the same diabetes individual in China.J Med Microbiol. 2021 Mar;70(3). doi: 10.1099/jmm.0.001316. Epub 2021 Jan 29. J Med Microbiol. 2021. PMID: 33528353
-
Genetic Characterization of Enterobacter hormaechei Co-Harboring bla NDM-1 and mcr-9 Causing Upper Respiratory Tract Infection.Infect Drug Resist. 2022 Aug 31;15:5035-5042. doi: 10.2147/IDR.S367073. eCollection 2022. Infect Drug Resist. 2022. PMID: 36068833 Free PMC article.
Cited by
-
Coexistence of Multidrug Resistance and Virulence in a Single Conjugative Plasmid from a Hypervirulent Klebsiella pneumoniae Isolate of Sequence Type 25.mSphere. 2022 Dec 21;7(6):e0047722. doi: 10.1128/msphere.00477-22. Epub 2022 Dec 6. mSphere. 2022. PMID: 36472445 Free PMC article.
-
The Co-occurrence of NDM-5, MCR-1, and FosA3-Encoding Plasmids Contributed to the Generation of Extensively Drug-Resistant Klebsiella pneumoniae.Front Microbiol. 2022 Jan 3;12:811263. doi: 10.3389/fmicb.2021.811263. eCollection 2021. Front Microbiol. 2022. PMID: 35046925 Free PMC article.
-
Co-occurrence of bla NDM-1 and mcr-9 in a Conjugative IncHI2/HI2A Plasmid From a Bloodstream Infection-Causing Carbapenem-Resistant Klebsiella pneumoniae.Front Microbiol. 2021 Nov 30;12:756201. doi: 10.3389/fmicb.2021.756201. eCollection 2021. Front Microbiol. 2021. PMID: 34956120 Free PMC article.
-
First Case Report of Detection of Multidrug-Resistant Enterobacter hormaechei in Clinical Sample from an Aborted Ruminant.Microorganisms. 2022 May 17;10(5):1036. doi: 10.3390/microorganisms10051036. Microorganisms. 2022. PMID: 35630478 Free PMC article.
-
Emergence of Neonatal Sepsis Caused by MCR-9- and NDM-1-Co-Producing Enterobacter hormaechei in China.Front Cell Infect Microbiol. 2022 May 6;12:879409. doi: 10.3389/fcimb.2022.879409. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35601097 Free PMC article.
References
-
- Bertrand S., Weill F., Cloeckaert A., Vrints M., Mairiaux E., Praud K., et al. (2006). Clonal emergence of extended-spectrum beta-lactamase (CTX-M-2)-producing Salmonella enterica serovar Virchow isolates with reduced susceptibilities to ciprofloxacin among poultry and humans in Belgium and France (2000 to 2003). J. Clin. Microbiol. 44 2897–2903. 10.1128/jcm.02549-05 - DOI - PMC - PubMed
-
- Börjesson S., Greko C., Myrenas M., Landén A., Nilsson O., Pedersen K. (2020). A link between the newly described colistin resistance gene mcr-9 and clinical Enterobacteriaceae isolates carrying bla from horses in Sweden. J. Glob. Antimicrob. Resist. 20 285–289. 10.1016/j.jgar.2019.08.007 - DOI - PubMed
LinkOut - more resources
Full Text Sources

