An Integrated Approach to Determine the Boundaries of the Azaphilone Pigment Biosynthetic Gene Cluster of Monascus ruber M7 Grown on Potato Dextrose Agar
- PMID: 34220766
- PMCID: PMC8241920
- DOI: 10.3389/fmicb.2021.680629
An Integrated Approach to Determine the Boundaries of the Azaphilone Pigment Biosynthetic Gene Cluster of Monascus ruber M7 Grown on Potato Dextrose Agar
Abstract
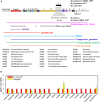
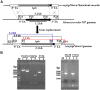
Monascus-type azaphilone pigments (MonAzPs) are produced in multi-thousand ton quantities each year and used as food colorants and nutraceuticals in East Asia. Several groups, including ours, described MonAzPs biosynthesis as a highly complex pathway with many branch points, affording more than 110 MonAzP congeners in a small group of fungi in the Eurotiales order. MonAzPs biosynthetic gene clusters (BGCs) are also very complex and mosaic-like, with some genes involved in more than one pathway, while other genes playing no apparent role in MonAzPs production. Due to this complexity, MonAzPs BGCs have been delimited differently in various fungi. Since most of these predictions rely primarily on bioinformatic analyses, it is possible that genes immediately outside the currently predicted BGC borders are also involved, especially those whose function cannot be predicted from sequence similarities alone. Conversely, some peripheral genes presumed to be part of the BGC may in fact lay outside the boundaries. This study uses a combination of computational and transcriptional analyses to predict the extent of the MonAzPs BGC in Monascus ruber M7. Gene knockouts and analysis of MonAzPs production of the mutants are then used to validate the prediction, revealing that the BGC consists of 16 genes, extending from mrpigA to mrpigP. We further predict that two strains of Talaromyces marneffei, ATCC 18224 and PM1, encode an orthologous but non-syntenic MonAzPs BGC with 14 genes. This work highlights the need to use comprehensive, integrated approaches for the more precise determination of secondary metabolite BGC boundaries.
Keywords: Monascus azaphilone pigment; comparative genomics; gene cluster boundary; gene knockout; transcription analysis.
Copyright © 2021 Liu, Zhong, Wang, Gao, Yang, Chen and Molnár.
Conflict of interest statement
IM has disclosed financial interests in Teva Pharmaceutical Works Ltd., Hungary and the University of Debrecen, Hungary that are unrelated to the subject of the research presented here. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Orange, red, yellow: biosynthesis of azaphilone pigments in Monascus fungi.Chem Sci. 2017 Jul 1;8(7):4917-4925. doi: 10.1039/c7sc00475c. Epub 2017 Apr 24. Chem Sci. 2017. PMID: 28959415 Free PMC article.
-
Enhancement of Monascus yellow pigments production by activating the cAMP signalling pathway in Monascus purpureus HJ11.Microb Cell Fact. 2020 Dec 7;19(1):224. doi: 10.1186/s12934-020-01486-y. Microb Cell Fact. 2020. PMID: 33287814 Free PMC article.
-
Nature and nurture: confluence of pathway determinism with metabolic and chemical serendipity diversifies Monascus azaphilone pigments.Nat Prod Rep. 2019 Apr 17;36(4):561-572. doi: 10.1039/c8np00060c. Nat Prod Rep. 2019. PMID: 30484470 Free PMC article. Review.
-
The regulation mechanisms of soluble starch and glycerol for production of azaphilone pigments in Monascus purpureus FAFU618 as revealed by comparative proteomic and transcriptional analyses.Food Res Int. 2018 Apr;106:626-635. doi: 10.1016/j.foodres.2018.01.037. Epub 2018 Feb 8. Food Res Int. 2018. PMID: 29579968
-
Azaphilone alkaloids: prospective source of natural food pigments.Appl Microbiol Biotechnol. 2022 Jan;106(2):469-484. doi: 10.1007/s00253-021-11729-6. Epub 2021 Dec 18. Appl Microbiol Biotechnol. 2022. PMID: 34921328 Review.
Cited by
-
Inactivation of mrpigH Gene in Monascus ruber M7 Results in Increased Monascus Pigments and Decreased Citrinin with mrpyrG Selection Marker.J Fungi (Basel). 2021 Dec 19;7(12):1094. doi: 10.3390/jof7121094. J Fungi (Basel). 2021. PMID: 34947076 Free PMC article.
References
-
- Balakrishnan B., Chen C.-C., Pan T.-M., Kwon H.-J. (2014a). Mpp7 controls regioselective Knoevenagel condensation during the biosynthesis of Monascus azaphilone pigments. Tetrahedron Lett. 55 1640–1643. 10.1016/j.tetlet.2014.01.090 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

