Genomic Analysis of the Endophytic Stenotrophomonas Strain 169 Reveals Features Related to Plant-Growth Promotion and Stress Tolerance
- PMID: 34220780
- PMCID: PMC8245107
- DOI: 10.3389/fmicb.2021.687463
Genomic Analysis of the Endophytic Stenotrophomonas Strain 169 Reveals Features Related to Plant-Growth Promotion and Stress Tolerance
Abstract
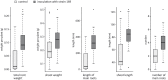
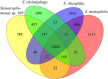
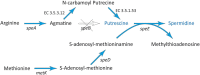
Plant-associated Stenotrophomonas isolates have great potential for plant growth promotion, especially under stress conditions, due to their ability to promote tolerance to abiotic stresses such as salinity or drought. The endophytic strain Stenotrophomonas sp. 169, isolated from a field-grown poplar, increased the growth of inoculated in vitro plants, with a particular effect on root development, and was able to stimulate the rooting of poplar cuttings in the greenhouse. The strain produced high amounts of the plant growth-stimulating hormone auxin under in vitro conditions. The comparison of the 16S rRNA gene sequences and the phylogenetic analysis of the core genomes showed a close relationship to Stenotrophomonas chelatiphaga and a clear separation from Stenotrophomonas maltophilia. Whole genome sequence analysis revealed functional genes potentially associated with attachment and plant colonization, growth promotion, and stress protection. In detail, an extensive set of genes for twitching motility, chemotaxis, flagella biosynthesis, and the ability to form biofilms, which are connected with host plant colonization, could be identified in the genome of strain 169. The production of indole-3-acetic acid and the presence of genes for auxin biosynthesis pathways and the spermidine pathway could explain the ability to promote plant growth. Furthermore, the genome contained genes encoding for features related to the production of different osmoprotective molecules and enzymes mediating the regulation of stress tolerance and the ability of bacteria to quickly adapt to changing environments. Overall, the results of physiological tests and genome analysis demonstrated the capability of endophytic strain 169 to promote plant growth. In contrast to related species, strain 169 can be considered non-pathogenic and suitable for biotechnology applications.
Keywords: auxin; endophytic bacteria; genome mining; phylogenomics; plant growth promotion; plant-microbe interaction.
Copyright © 2021 Ulrich, Kube, Becker, Schneck and Ulrich.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees.Appl Environ Microbiol. 2009 Feb;75(3):748-57. doi: 10.1128/AEM.02239-08. Epub 2008 Dec 5. Appl Environ Microbiol. 2009. PMID: 19060168 Free PMC article.
-
A comprehensive comparative genomic analysis revealed that plant growth promoting traits are ubiquitous in strains of Stenotrophomonas.Front Microbiol. 2024 May 16;15:1395477. doi: 10.3389/fmicb.2024.1395477. eCollection 2024. Front Microbiol. 2024. PMID: 38817968 Free PMC article.
-
Genome-Guided Insights into the Plant Growth Promotion Capabilities of the Physiologically Versatile Bacillus aryabhattai Strain AB211.Front Microbiol. 2017 Mar 21;8:411. doi: 10.3389/fmicb.2017.00411. eCollection 2017. Front Microbiol. 2017. PMID: 28377746 Free PMC article.
-
Drought tolerance improvement in plants: an endophytic bacterial approach.Appl Microbiol Biotechnol. 2019 Sep;103(18):7385-7397. doi: 10.1007/s00253-019-10045-4. Epub 2019 Aug 2. Appl Microbiol Biotechnol. 2019. PMID: 31375881 Review.
-
Induction of abiotic stress tolerance in plants by endophytic microbes.Lett Appl Microbiol. 2018 Apr;66(4):268-276. doi: 10.1111/lam.12855. Epub 2018 Feb 26. Lett Appl Microbiol. 2018. PMID: 29359344 Review.
Cited by
-
Multiple Chitin- or Avirulent Strain-Triggered Immunity Induces Microbiome Reassembly in Rice.Microorganisms. 2024 Jun 28;12(7):1323. doi: 10.3390/microorganisms12071323. Microorganisms. 2024. PMID: 39065092 Free PMC article.
-
Stenotrophomonas maltophilia promotes wheat growth by enhancing nutrient assimilation and rhizosphere microbiota modulation.Front Bioeng Biotechnol. 2025 Apr 17;13:1563670. doi: 10.3389/fbioe.2025.1563670. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40313642 Free PMC article.
-
Endophytic Bacterium Paenibacillus illinoisensis Y11 Promotes Duckweed Growth and Enhances Cadmium Pollution Remediation.Curr Microbiol. 2025 Aug 12;82(10):447. doi: 10.1007/s00284-025-04451-3. Curr Microbiol. 2025. PMID: 40797047
-
Consumption of N2O by Flavobacterium azooxidireducens sp. nov. Isolated from Decomposing Leaf Litter of Phragmites australis (Cav.).Microorganisms. 2022 Nov 21;10(11):2304. doi: 10.3390/microorganisms10112304. Microorganisms. 2022. PMID: 36422374 Free PMC article.
-
Characterization of three Stenotrophomonas strains isolated from different ecosystems and proposal of Stenotrophomonas mori sp. nov. and Stenotrophomonas lacuserhaii sp. nov.Front Microbiol. 2022 Dec 15;13:1056762. doi: 10.3389/fmicb.2022.1056762. eCollection 2022. Front Microbiol. 2022. PMID: 36590414 Free PMC article.
References
-
- Abdelaziz S., Hemeda N. F., Belal E. E., Elshahawy R. (2018). Efficacy of facultative oligotrophic bacterial strains as plant growth-promoting rhizobacteria (pgpr) and their potency against two pathogenic fungi causing damping-off disease. Appl. Microbiol. Open Access 4:153. 10.4172/2471-9315.1000153 - DOI
LinkOut - more resources
Full Text Sources

