Recent Advances in Molecular Biology of Human Bocavirus 1 and Its Applications
- PMID: 34220786
- PMCID: PMC8242256
- DOI: 10.3389/fmicb.2021.696604
Recent Advances in Molecular Biology of Human Bocavirus 1 and Its Applications
Abstract
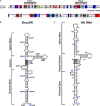
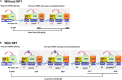
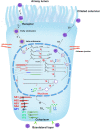
Human bocavirus 1 (HBoV1) was discovered in human nasopharyngeal specimens in 2005. It is an autonomous human parvovirus and causes acute respiratory tract infections in young children. HBoV1 infects well differentiated or polarized human airway epithelial cells in vitro. Unique among all parvoviruses, HBoV1 expresses 6 non-structural proteins, NS1, NS1-70, NS2, NS3, NS4, and NP1, and a viral non-coding RNA (BocaSR), and three structural proteins VP1, VP2, and VP3. The BocaSR is the first identified RNA polymerase III (Pol III) transcribed viral non-coding RNA in small DNA viruses. It plays an important role in regulation of viral gene expression and a direct role in viral DNA replication in the nucleus. HBoV1 genome replication in the polarized/non-dividing airway epithelial cells depends on the DNA damage and DNA repair pathways and involves error-free Y-family DNA repair DNA polymerase (Pol) η and Pol κ. Importantly, HBoV1 is a helper virus for the replication of dependoparvovirus, adeno-associated virus (AAV), in polarized human airway epithelial cells, and HBoV1 gene products support wild-type AAV replication and recombinant AAV (rAAV) production in human embryonic kidney (HEK) 293 cells. More importantly, the HBoV1 capsid is able to pseudopackage an rAAV2 or rHBoV1 genome, producing the rAAV2/HBoV1 or rHBoV1 vector. The HBoV1 capsid based rAAV vector has a high tropism for human airway epithelia. A deeper understanding in HBoV1 replication and gene expression will help find a better way to produce the rAAV vector and to increase the efficacy of gene delivery using the rAAV2/HBoV1 or rHBoV1 vector, in particular, to human airways. This review summarizes the recent advances in gene expression and replication of HBoV1, as well as the use of HBoV1 as a parvoviral vector for gene delivery.
Keywords: gene expression; human bocavirus; parvovirus; replication; viral vector.
Copyright © 2021 Shao, Shen, Wang and Qiu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
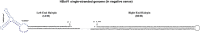


Similar articles
-
Parvovirus Expresses a Small Noncoding RNA That Plays an Essential Role in Virus Replication.J Virol. 2017 Mar 29;91(8):e02375-16. doi: 10.1128/JVI.02375-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28122984 Free PMC article.
-
Human Bocavirus 1 Is a Novel Helper for Adeno-associated Virus Replication.J Virol. 2017 Aug 24;91(18):e00710-17. doi: 10.1128/JVI.00710-17. Print 2017 Sep 15. J Virol. 2017. PMID: 28659483 Free PMC article.
-
Nonstructural Protein NP1 of Human Bocavirus 1 Plays a Critical Role in the Expression of Viral Capsid Proteins.J Virol. 2016 Apr 14;90(9):4658-4669. doi: 10.1128/JVI.02964-15. Print 2016 May. J Virol. 2016. PMID: 26912614 Free PMC article.
-
[Progress on development and research of human bocavirus 1].Bing Du Xue Bao. 2014 Jan;30(1):103-8. Bing Du Xue Bao. 2014. PMID: 24772907 Review. Chinese.
-
Human bocavirus: Current knowledge and future challenges.World J Gastroenterol. 2016 Oct 21;22(39):8684-8697. doi: 10.3748/wjg.v22.i39.8684. World J Gastroenterol. 2016. PMID: 27818586 Free PMC article. Review.
Cited by
-
Neurological Impact of Respiratory Viruses: Insights into Glial Cell Responses in the Central Nervous System.Microorganisms. 2024 Aug 20;12(8):1713. doi: 10.3390/microorganisms12081713. Microorganisms. 2024. PMID: 39203555 Free PMC article. Review.
-
Human Bocavirus 1 NP1 acts as an ssDNA-binding protein to help AAV2 DNA replication and cooperates with RPA to regulate AAV2 capsid expression.J Virol. 2024 Mar 19;98(3):e0151523. doi: 10.1128/jvi.01515-23. Epub 2024 Feb 7. J Virol. 2024. PMID: 38323812 Free PMC article.
-
For better or worse: crosstalk of parvovirus and host DNA damage response.Front Immunol. 2024 Feb 23;15:1324531. doi: 10.3389/fimmu.2024.1324531. eCollection 2024. Front Immunol. 2024. PMID: 38464523 Free PMC article. Review.
-
Genetic characteristics of human bocavirus in children with acute respiratory tract infections during 2023 in Beijing, China.Virol J. 2025 Jun 30;22(1):212. doi: 10.1186/s12985-025-02846-z. Virol J. 2025. PMID: 40588726 Free PMC article.
-
Prevalence and molecular characterization of human bocavirus-1 in children and adults with influenza-like illness from Kunming, Southwest China.Microbiol Spectr. 2025 Jan 7;13(1):e0156424. doi: 10.1128/spectrum.01564-24. Epub 2024 Dec 11. Microbiol Spectr. 2025. PMID: 39660928 Free PMC article.
References
-
- Astell C. R., Thomson M., Chow M. B., Ward D. C. (1983). Structure and replication of minute virus of mice DNA. Cold Spring Harb. Symp. Quant. Biol. 47 Pt 2 751–762. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

