Immune Tolerance-Adjusted Personalized Immunogenicity Prediction for Pompe Disease
- PMID: 34220802
- PMCID: PMC8242953
- DOI: 10.3389/fimmu.2021.636731
Immune Tolerance-Adjusted Personalized Immunogenicity Prediction for Pompe Disease
Abstract
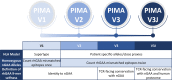
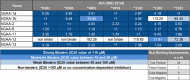
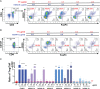
Infantile-onset Pompe disease (IOPD) is a glycogen storage disease caused by a deficiency of acid alpha-glucosidase (GAA). Treatment with recombinant human GAA (rhGAA, alglucosidase alfa) enzyme replacement therapy (ERT) significantly improves clinical outcomes; however, many IOPD children treated with rhGAA develop anti-drug antibodies (ADA) that render the therapy ineffective. Antibodies to rhGAA are driven by T cell responses to sequences in rhGAA that differ from the individuals' native GAA (nGAA). The goal of this study was to develop a tool for personalized immunogenicity risk assessment (PIMA) that quantifies T cell epitopes that differ between nGAA and rhGAA using information about an individual's native GAA gene and their HLA DR haplotype, and to use this information to predict the risk of developing ADA. Four versions of PIMA have been developed. They use EpiMatrix, a computational tool for T cell epitope identification, combined with an HLA-restricted epitope-specific scoring feature (iTEM), to assess ADA risk. One version of PIMA also integrates JanusMatrix, a Treg epitope prediction tool to identify putative immunomodulatory (regulatory) T cell epitopes in self-proteins. Using the JanusMatrix-adjusted version of PIMA in a logistic regression model with data from 48 cross-reactive immunological material (CRIM)-positive IOPD subjects, those with scores greater than 10 were 4-fold more likely to develop ADA (p<0.03) than those that had scores less than 10. We also confirmed the hypothesis that some GAA epitopes are immunomodulatory. Twenty-one epitopes were tested, of which four were determined to have an immunomodulatory effect on T effector response in vitro. The implementation of PIMA V3J on a secure-access website would allow clinicians to input the individual HLA DR haplotype of their IOPD patient and the GAA pathogenic variants associated with each GAA allele to calculate the patient's relative risk of developing ADA, enhancing clinical decision-making prior to initiating treatment with ERT. A better understanding of immunogenicity risk will allow the implementation of targeted immunomodulatory approaches in ERT-naïve settings, especially in CRIM-positive patients, which may in turn improve the overall clinical outcomes by minimizing the development of ADA. The PIMA approach may also be useful for other types of enzyme or factor replacement therapies.
Keywords: Pompe Disease (glycogen storage disease type II); Tregitope; acid alpha-glucosidase (GAA); anti-drug antibodies (ADA); cross-reactive immunological material (CRIM); enzyme replacement therapy (ERT); immune tolerance induction (ITI); personalized immunogenicity assessment (PIMA).
Copyright © 2021 De Groot, Desai, Lelias, Miah, Terry, Khan, Li, Yi, Ardito, Martin and Kishnani.
Conflict of interest statement
ADG and WDM are senior officers and shareholders, and SM, FET, SK, MA, and SL are employees of EpiVax, Inc., a company specializing in immunoinformatic analysis. EpiVax, Inc. own patents to technologies utilized by associated authors in the research reported here. AKD has received grant support from Sanofi Genzyme and the lysosomal disease network. PSK has received research/grant support from Sanofi Genzyme, Valerion Therapeutics, and Amicus Therapeutics. PSK has received consulting fees and honoraria from Sanofi Genzyme, Amicus Therapeutics, Maze Therapeutics, JCR Pharmaceutical and Asklepios Biopharmaceutical, Inc. (AskBio). PSK is a member of the Pompe and Gaucher Disease Registry Advisory Board for Sanofi Genzyme, Amicus Therapeutics, and Baebies. PSK has equity in Asklepios Biopharmaceutical, Inc. (AskBio), which is developing gene therapy for Pompe disease and Maze Therapeutics, which is developing small molecule in Pompe disease. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
HLA- and genotype-based risk assessment model to identify infantile onset pompe disease patients at high-risk of developing significant anti-drug antibodies (ADA).Clin Immunol. 2019 Mar;200:66-70. doi: 10.1016/j.clim.2019.01.009. Epub 2019 Jan 31. Clin Immunol. 2019. PMID: 30711607 Free PMC article.
-
Teaching tolerance: New approaches to enzyme replacement therapy for Pompe disease.Hum Vaccin Immunother. 2012 Oct;8(10):1459-64. doi: 10.4161/hv.21405. Epub 2012 Oct 1. Hum Vaccin Immunother. 2012. PMID: 23095864 Free PMC article.
-
Optimizing treatment outcomes: immune tolerance induction in Pompe disease patients undergoing enzyme replacement therapy.Front Immunol. 2024 Apr 23;15:1336599. doi: 10.3389/fimmu.2024.1336599. eCollection 2024. Front Immunol. 2024. PMID: 38715621 Free PMC article. Clinical Trial.
-
Gene Therapy for Pompe Disease: The Time is now.Hum Gene Ther. 2019 Oct;30(10):1245-1262. doi: 10.1089/hum.2019.109. Epub 2019 Sep 9. Hum Gene Ther. 2019. PMID: 31298581 Review.
-
Pompe disease: early diagnosis and early treatment make a difference.Pediatr Neonatol. 2013 Aug;54(4):219-27. doi: 10.1016/j.pedneo.2013.03.009. Epub 2013 Apr 28. Pediatr Neonatol. 2013. PMID: 23632029 Review.
Cited by
-
What's new and what's next for gene therapy in Pompe disease?Expert Opin Biol Ther. 2022 Sep;22(9):1117-1135. doi: 10.1080/14712598.2022.2067476. Epub 2022 Apr 27. Expert Opin Biol Ther. 2022. PMID: 35428407 Free PMC article.
-
In silico methods for immunogenicity risk assessment and human homology screening for therapeutic antibodies.MAbs. 2024 Jan-Dec;16(1):2333729. doi: 10.1080/19420862.2024.2333729. Epub 2024 Mar 27. MAbs. 2024. PMID: 38536724 Free PMC article.
-
Case Report: Identification of Compound Heterozygous Mutations in a Patient With Late-Onset Glycogen Storage Disease Type II (Pompe Disease).Front Neurol. 2022 Mar 21;13:839263. doi: 10.3389/fneur.2022.839263. eCollection 2022. Front Neurol. 2022. PMID: 35386406 Free PMC article.
-
Regulatory T cell epitope content in human antibodies decreases during maturation.Front Immunol. 2025 Apr 17;16:1535826. doi: 10.3389/fimmu.2025.1535826. eCollection 2025. Front Immunol. 2025. PMID: 40313951 Free PMC article.
-
Antibodies against recombinant human alpha-glucosidase do not seem to affect clinical outcome in childhood onset Pompe disease.Orphanet J Rare Dis. 2022 Feb 2;17(1):31. doi: 10.1186/s13023-022-02175-2. Orphanet J Rare Dis. 2022. PMID: 35109913 Free PMC article.
References
-
- Scriver C, Beaudet A, Sly W, Valle D. Glycogen Storage Disease Type II: Acid Alpha-Glucosidase (Acid Maltase) Deficiency. In: Hirshhorn R, editor. The Metabolic and Molecular Bases of Inherited Disease. A. Reuser New York: McGraw-Hill; (2001).
-
- Bali DS, Goldstein JL, Banugaria S, Dai J, Mackey J, Rehder C, et al. . Predicting Cross-Reactive Immunological Material (CRIM) Status in Pompe Disease Using GAA Mutations: Lessons Learned From 10 Years of Clinical Laboratory Testing Experience. Am J Med Genet Part C Semin Med Genet (2012) 160C:40–9. 10.1002/ajmg.c.31319 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

