Significance of PD1 Alternative Splicing in Celiac Disease as a Novel Source for Diagnostic and Therapeutic Target
- PMID: 34220824
- PMCID: PMC8242946
- DOI: 10.3389/fimmu.2021.678400
Significance of PD1 Alternative Splicing in Celiac Disease as a Novel Source for Diagnostic and Therapeutic Target
Abstract
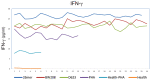
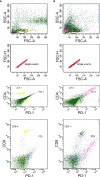
Background: We have focused on the alteration of the PD-1/PD-L1 pathway in celiac disease and discussed the roles of the PD1 pathway in regulating the immune response. We explored the idea that the altered mRNA splicing process in key regulatory proteins could represent a novel source to identify diagnostic, prognostic, and therapeutic targets in celiac disease.
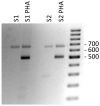
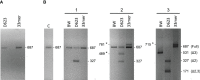
Methods: We characterized the PD1 mRNA variants' profile in CD patients and in response to gluten peptides' incubation after in vitro experiments. Total RNA from whole blood was isolated, and the coding region of the human PD-1 mRNA was amplified by cDNA PCR.
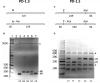
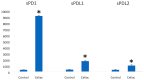
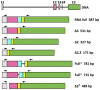
Results: PCR amplification of the human PD-1 coding sequence revealed an association between the over-expression of the sPD-1 protein and the PD-1Δex3 transcript in celiac disease. Thus, we have found three novel alternative spliced isoforms, two of which result in a truncated protein and the other isoform with a loss of 14 aa of exon 2 and complete exon 3 (Δ3) which could encode a new soluble form of PD1 (sPD-1).
Conclusions: Our study provides evidence that dietary gluten can modulate processes required for cell homeostasis through the splicing of pre-mRNAs encoding key regulatory proteins, which represents an adaptive mechanism in response to different nutritional conditions.
Keywords: PD1/PDL; alternative splicing; celiac disease; gluten peptides; immune checkpoint.
Copyright © 2021 Ponce de León, Lorite, López-Casado, Barro, Palomeque and Torres.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Targeting alternative splicing as a new cancer immunotherapy-phosphorylation of serine arginine-rich splicing factor (SRSF1) by SR protein kinase 1 (SRPK1) regulates alternative splicing of PD1 to generate a soluble antagonistic isoform that prevents T cell exhaustion.Cancer Immunol Immunother. 2023 Dec;72(12):4001-4014. doi: 10.1007/s00262-023-03534-z. Epub 2023 Nov 16. Cancer Immunol Immunother. 2023. PMID: 37973660 Free PMC article.
-
PD1/PD-L1 Expressions in Plasmablastic Lymphoma with Clinicopathological Correlation.Ann Clin Lab Sci. 2021 Mar;51(2):174-181. Ann Clin Lab Sci. 2021. PMID: 33941556
-
An Extended PD-L2 Cytoplasmic Domain Results From Alternative Splicing in NSCLC Cells.J Immunother. 2022 Nov-Dec 01;45(9):379-388. doi: 10.1097/CJI.0000000000000439. Epub 2022 Aug 30. J Immunother. 2022. PMID: 36036966
-
Study and analysis of antitumor resistance mechanism of PD1/PD-L1 immune checkpoint blocker.Cancer Med. 2020 Nov;9(21):8086-8121. doi: 10.1002/cam4.3410. Epub 2020 Sep 2. Cancer Med. 2020. PMID: 32875727 Free PMC article. Review.
-
The Roles of microRNAs in Regulating the Expression of PD-1/PD-L1 Immune Checkpoint.Int J Mol Sci. 2017 Nov 27;18(12):2540. doi: 10.3390/ijms18122540. Int J Mol Sci. 2017. PMID: 29186904 Free PMC article. Review.
Cited by
-
Role of sPD-1 and sPD-Ls in the pathogenesis of connective tissue disease.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2023 Mar 28;48(3):444-454. doi: 10.11817/j.issn.1672-7347.2023.220263. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2023. PMID: 37164928 Free PMC article. Chinese, English.
-
Expression of Elafin and CD200 as Immune Checkpoint Molecules Involved in Celiac Disease.Int J Mol Sci. 2024 Jan 10;25(2):852. doi: 10.3390/ijms25020852. Int J Mol Sci. 2024. PMID: 38255930 Free PMC article.
-
Comprehensive characterization of the alternative splicing landscape in ovarian cancer reveals novel events associated with tumor-immune microenvironment.Biosci Rep. 2022 Feb 25;42(2):BSR20212090. doi: 10.1042/BSR20212090. Biosci Rep. 2022. PMID: 35137909 Free PMC article.
-
A splicing isoform of PD-1 promotes tumor progression as a potential immune checkpoint.Nat Commun. 2024 Oct 23;15(1):9114. doi: 10.1038/s41467-024-53561-2. Nat Commun. 2024. PMID: 39438489 Free PMC article.
-
Identification of Alternative Splicing-Related Genes CYB561 and FOLH1 in the Tumor-Immune Microenvironment for Endometrial Cancer Based on TCGA Data Analysis.Front Genet. 2022 Jun 28;13:770569. doi: 10.3389/fgene.2022.770569. eCollection 2022. Front Genet. 2022. PMID: 35836577 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

