lme4GS: An R-Package for Genomic Selection
- PMID: 34220954
- PMCID: PMC8250143
- DOI: 10.3389/fgene.2021.680569
lme4GS: An R-Package for Genomic Selection
Abstract
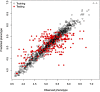
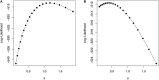
Genomic selection (GS) is a technology used for genetic improvement, and it has many advantages over phenotype-based selection. There are several statistical models that adequately approach the statistical challenges in GS, such as in linear mixed models (LMMs). An active area of research is the development of software for fitting LMMs mainly used to make genome-based predictions. The lme4 is the standard package for fitting linear and generalized LMMs in the R-package, but its use for genetic analysis is limited because it does not allow the correlation between individuals or groups of individuals to be defined. This article describes the new lme4GS package for R, which is focused on fitting LMMs with covariance structures defined by the user, bandwidth selection, and genomic prediction. The new package is focused on genomic prediction of the models used in GS and can fit LMMs using different variance-covariance matrices. Several examples of GS models are presented using this package as well as the analysis using real data.
Keywords: genomic prediction; genomic selection; kernel; linear mixed model; lme4.
Copyright © 2021 Caamal-Pat, Pérez-Rodríguez, Crossa, Velasco-Cruz, Pérez-Elizalde and Vázquez-Peña.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Acosta-Pech R., Crossa J., de los Campos G., Teyssèdre S., Claustres B., Pérez-Elizalde S., et al. (2017). Genomic models with genotype x environment interaction for predicting hybrid performance: an application in maize hybrids. Theoretical and Applied Genetics 130 1431–1440. 10.1007/s00122-017-2898-0 - DOI - PubMed
-
- Bates D., Mächler M., Bolker B. M., Walker S. C. (2015). Fitting linear mixed-effects models using lme4. Journal of Statistical Software 67 1–48. 10.18637/jss.v067.i01 - DOI
-
- Bernardo R., Yu J. (2007). Prospects for genome wide selection for quantitative traits in maize. Crop Science 47 1082–1090. 10.2135/cropsci2006.11.0690 - DOI
LinkOut - more resources
Full Text Sources