CircRNA expression profiles and circRNA-miRNA-mRNA crosstalk in allergic rhinitis
- PMID: 34221216
- PMCID: PMC8233375
- DOI: 10.1016/j.waojou.2021.100548
CircRNA expression profiles and circRNA-miRNA-mRNA crosstalk in allergic rhinitis
Abstract
Background: Circular RNAs (circRNAs) are involved in inflammation; however, their role in allergic rhinitis (AR) remains unclear. In this study, we analyzed circRNA expression and identified a circRNA-miRNA-mRNA network through which circRNAs regulate AR pathogenesis.
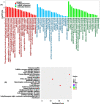
Methods: We analyzed circRNA, miRNA, and mRNA expression profiles in the nasal mucosa by high-throughput sequencing (HTS), using a fold-change >1.5 and p-value < 0.05 to pinpoint significantly differentially expressed (DE) circRNAs, miRNAs, and mRNAs in AR. A DEcircRNA-DEmiRNA-DEmRNA crosstalk network was then constructed using bioinformatics and statistical analysis. Gene ontology and Kyoto encyclopedia of genes and genomes pathway analyses were performed to identify the biological terms enriched in the network; whereas RT-PCR and Sanger sequencing were used to confirm the circRNAs.
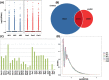
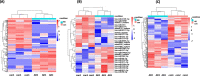
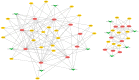
Results: A total of 264 DEcircRNAs were identified by HTS, including 120 upregulated and 144 downregulated in AR compared to controls. A DEcircRNA-DEmiRNA-DEmRNA crosstalk network was constructed with 17 miRNAs, 11 circRNAs, 29 mRNAs, and 64 interaction pairs. These genes were involved in the Wnt signaling pathway, TNF biosynthesis, inflammatory responses, the PI3K-Akt signaling pathway, and Toll-like receptors. Of the 11 DEcircRNAs, hsa_circ_0008668 and circTRIQK were upregulated, whereas hsa_circ_0029853 and circRNA_01002 were downregulated in AR tissues. Sanger sequencing confirmed the back-splicing junctions of these circRNAs.
Conclusions: We constructed a novel DEcircRNA-DEmiRNA-DEmRNA network for AR that provides a basis for future studies to investigate its underlying molecular mechanisms.
Keywords: Allergic rhinitis; Circular RNA; Nasal mucosa; mRNA; microRNA.
© 2021 The Author(s).
Figures



References
-
- Shaoqing Y., Ruxin Z., Guojun L. Microarray analysis of differentially expressed microRNAs in allergic rhinitis. Am J Rhinol Allergy. 2011;25:e242–e246. - PubMed
-
- Ma Z., Teng Y., Liu X. Identification and functional profiling of differentially expressed long non-coding RNAs in nasal mucosa with allergic rhinitis. Tohoku J Exp Med. 2017;242:143–150. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

