Comprehensive transcriptome analysis of peripheral blood unravels key lncRNAs implicated in ABPA and asthma
- PMID: 34221710
- PMCID: PMC8236232
- DOI: 10.7717/peerj.11453
Comprehensive transcriptome analysis of peripheral blood unravels key lncRNAs implicated in ABPA and asthma
Abstract
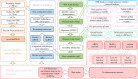
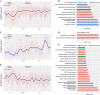
Allergic bronchopulmonary aspergillosis (ABPA) is a complex hypersensitivity lung disease caused by a fungus known as Aspergillus fumigatus. It complicates and aggravates asthma. Despite their potential associations, the underlying mechanisms of asthma developing into ABPA remain obscure. Here we performed an integrative transcriptome analysis based on three types of human peripheral blood, which derived from ABPA patients, asthmatic patients and health controls, aiming to identify crucial lncRNAs implicated in ABPA and asthma. Initially, a high-confidence dataset of lncRNAs was identified using a stringent filtering pipeline. A comparative mutational analysis revealed no significant difference among these samples. Differential expression analysis disclosed several immune-related mRNAs and lncRNAs differentially expressed in ABPA and asthma. For each disease, three sub-networks were established using differential network analysis. Many key lncRNAs implicated in ABPA and asthma were identified, respectively, i.e., AL139423.1-201, AC106028.4-201, HNRNPUL1-210, PUF60-218 and SREBF1-208. Our analysis indicated that these lncRNAs exhibits in the loss-of-function networks, and the expression of which were repressed in the occurrences of both diseases, implying their important roles in the immune-related processes in response to the occurrence of both diseases. Above all, our analysis proposed a new point of view to explore the relationship between ABPA and asthma, which might provide new clues to unveil the pathogenic mechanisms for both diseases.
Keywords: ABPA; Asthma; Long noncoding RNAs; Network analysis; RNA sequencing; Transcriptome analysis.
© 2021 Huang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Transcriptome analysis of peripheral whole blood identifies crucial lncRNAs implicated in childhood asthma.BMC Med Genomics. 2020 Sep 18;13(1):136. doi: 10.1186/s12920-020-00785-y. BMC Med Genomics. 2020. PMID: 32948203 Free PMC article.
-
Induced sputum eosinophils and neutrophils and bronchiectasis severity in allergic bronchopulmonary aspergillosis.Eur Respir J. 2000 Dec;16(6):1095-101. doi: 10.1034/j.1399-3003.2000.16f13.x. Eur Respir J. 2000. PMID: 11292112
-
Aspergillus-associated hypersensitivity respiratory disorders.Indian J Chest Dis Allied Sci. 2008 Jan-Mar;50(1):117-28. Indian J Chest Dis Allied Sci. 2008. PMID: 18610696 Review.
-
An asthmatic child with allergic bronchopulmonary aspergillosis (ABPA).Turk J Pediatr. 2018;60(4):446-452. doi: 10.24953/turkjped.2018.04.017. Turk J Pediatr. 2018. PMID: 30859774
-
Allergic bronchopulmonary aspergillosis and related allergic syndromes.Semin Respir Crit Care Med. 2011 Dec;32(6):682-92. doi: 10.1055/s-0031-1295716. Epub 2011 Dec 13. Semin Respir Crit Care Med. 2011. PMID: 22167396 Review.
References
-
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

