Prognostic Value of Eight-Gene Signature in Head and Neck Squamous Carcinoma
- PMID: 34221975
- PMCID: PMC8252784
- DOI: 10.3389/fonc.2021.657002
Prognostic Value of Eight-Gene Signature in Head and Neck Squamous Carcinoma
Abstract
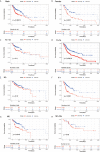
Head and neck cancer (HNC) is the fifth most common cancer worldwide. In this study, we performed an integrative analysis of the discovery set and established an eight-gene signature for the prediction of prognosis in patients with head and neck squamous cell carcinoma (HNSCC). Univariate Cox analysis was used to identify prognosis-related genes (with P < 0.05) in the GSE41613, GSE65858, and TCGA-HNSC RNA-Seq datasets after data collection. We performed LASSO Cox regression analysis and identified eight genes (CBX3, GNA12, P4HA1, PLAU, PPL, RAB25, EPHX3, and HLF) with non-zero regression coefficients in TCGA-HNSC datasets. Survival analysis revealed that the overall survival (OS) of GSE41613 and GSE65858 datasets and the progression-free survival(DFS)of GSE27020 and GSE42743 datasets in the low-risk group exhibited better survival outcomes compared with the high-risk group. To verify that the eight-mRNA prognostic model was independent of other clinical features, KM survival analysis of the specific subtypes with different clinical characteristics was performed. Univariate and multivariate Cox regression analyses were used to identify three independent prognostic factors to construct a prognostic nomogram. Finally, the GSVA algorithm identified six pathways that were activated in the intersection of the TCGA-HNSC, GSE65858, and GSE41613 datasets, including early estrogen response, cholesterol homeostasis, oxidative phosphorylation, fatty acid metabolism, bile acid metabolism, and Kras signaling. However, the epithelial-mesenchymal transition pathway was inhibited at the intersection of the three datasets. In conclusion, the eight-gene prognostic signature proved to be a useful tool in the prognostic evaluation and facilitate personalized treatment of HNSCC patients.
Keywords: Cox regression; GEO; LASSO; TCGA; head and neck squamous carcinoma; prognostic signature.
Copyright © 2021 Liu, Su, Ma, Chen, Wang, Che and Heng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources
Miscellaneous

