Characterisation of Bacteriophage-Encoded Depolymerases Selective for Key Klebsiella pneumoniae Capsular Exopolysaccharides
- PMID: 34222050
- PMCID: PMC8253255
- DOI: 10.3389/fcimb.2021.686090
Characterisation of Bacteriophage-Encoded Depolymerases Selective for Key Klebsiella pneumoniae Capsular Exopolysaccharides
Abstract
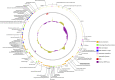
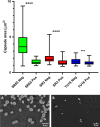
Capsular polysaccharides enable clinically important clones of Klebsiella pneumoniae to cause severe systemic infections in susceptible hosts. Phage-encoded capsule depolymerases have the potential to provide an alternative treatment paradigm in patients when multiple drug resistance has eroded the efficacy of conventional antibiotic chemotherapy. An investigation of 164 K. pneumoniae from intensive care patients in Thailand revealed a large number of distinct K types in low abundance but four (K2, K51, K1, K10) with a frequency of at least 5%. To identify depolymerases with the capacity to degrade capsules associated with these common K-types, 62 lytic phage were isolated from Thai hospital sewage water using K1, K2 and K51 isolates as hosts; phage plaques, without exception, displayed halos indicative of the presence of capsule-degrading enzymes. Phage genomes ranged in size from 41-348 kb with between 50 and 535 predicted coding sequences (CDSs). Using a custom phage protein database we were successful in applying annotation to 30 - 70% (mean = 58%) of these CDSs. The largest genomes, of so-called jumbo phage, carried multiple tRNAs as well as CRISPR repeat and spacer sequences. One of the smaller phage genomes was found to contain a putative Cas type 1E gene, indicating a history of host DNA acquisition in these obligate lytic phage. Whole-genome sequencing (WGS) indicated that some phage displayed an extended host range due to the presence of multiple depolymerase genes; in total, 42 candidate depolymerase genes were identified with up to eight in a single genome. Seven distinct virions were selected for further investigation on the basis of host range, phage morphology and WGS. Candidate genes for K1, K2 and K51 depolymerases were expressed and purified as his6-tagged soluble protein and enzymatic activity demonstrated against K. pneumoniae capsular polysaccharides by gel electrophoresis and Anton-Paar rolling ball viscometry. Depolymerases completely removed the capsule in K-type-specific fashion from K. pneumoniae cells. We conclude that broad-host range phage carry multiple enzymes, each with the capacity to degrade a single K-type, and any future use of these enzymes as therapeutic agents will require enzyme cocktails for utility against a range of K. pneumoniae infections.
Keywords: Klebsiella pneumoniae; alternative antibacterial therapy; bacteriophage; capsular polysaccharide; capsule depolymerase; jumbo phage; whole-genome sequencing (WGS).
Copyright © 2021 Blundell-Hunter, Enright, Negus, Dorman, Beecham, Pickard, Wintachai, Voravuthikunchai, Thomson and Taylor.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

