Wastewater Sample Site Selection to Estimate Geographically Resolved Community Prevalence of COVID-19: A Sampling Protocol Perspective
- PMID: 34222738
- PMCID: PMC8240399
- DOI: 10.1029/2021GH000420
Wastewater Sample Site Selection to Estimate Geographically Resolved Community Prevalence of COVID-19: A Sampling Protocol Perspective
Abstract
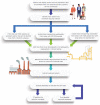
Wastewater monitoring for virus infections within communities can complement conventional clinical surveillance. Currently, most SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) clinical testing is voluntary and inconsistently available, except for a few occupational and educational settings, and therefore likely underrepresents actual population prevalence. Randomized testing on a regular basis to estimate accurate population-level infection rates is prohibitively costly and is hampered by a range of limitations and barriers associated with participation in clinical research. In comparison, community-level fecal monitoring can be performed through wastewater surveillance to effectively surveil communities. However, epidemiologically defined protocols for wastewater sample site selection are lacking. Herein, we describe methods for developing a geographically resolved population-level wastewater sampling approach in Jefferson County, Kentucky, and present preliminary results. Utilizing this site selection protocol, samples (n = 237) were collected from 17 wastewater catchment areas, September 8 to October 30, 2020 from one to four times per week in each area and compared to concurrent clinical data aggregated to wastewater catchment areas and county level. SARS-CoV-2 RNA was consistently present in wastewater during the studied period, and varied by area. Data obtained using the site selection protocol showed variation in geographically resolved wastewater SARS-CoV-2 RNA concentration compared to clinical rates. These findings highlight the importance of neighborhood-equivalent spatial scales and provide a promising approach for viral epidemic surveillance, thus better guiding spatially targeted public health mitigation strategies.
Keywords: COVID‐19; GIS; SARS‐CoV‐2; environmental surveillance; epidemiology; wastewater.
© 2021. The Authors. GeoHealth published by Wiley Periodicals LLC on behalf of American Geophysical Union.
Conflict of interest statement
The authors declare no conflicts of interest relevant to this study.
Figures


References
-
- Ahmed, W. , Angel, N. , Edson, J. , Bibby, K. , Bivins, A. , O'Brien, J. W. , et al. (2020). First confirmed detection of SARS‐CoV‐2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID‐19 in the community. Science of the Total Environment, 728, 138764. 10.1016/j.scitotenv.2020.138764 - DOI - PMC - PubMed
-
- Bisseux, M. , Debroas, D. , Mirand, A. , Archimbaud, C. , Peigue‐Lafeuille, H. , Bailly, J.‐L. , & Henquell, C. (2020). Monitoring of enterovirus diversity in wastewater by ultra‐deep sequencing: An effective complementary tool for clinical enterovirus surveillance. Water Research, 169, 115246. 10.1016/j.watres.2019.115246 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
