Exploring the evolution and epidemiology of European CC1-MRSA-IV: tracking a multidrug-resistant community-associated meticillin-resistant Staphylococcus aureus clone
- PMID: 34223815
- PMCID: PMC8477393
- DOI: 10.1099/mgen.0.000601
Exploring the evolution and epidemiology of European CC1-MRSA-IV: tracking a multidrug-resistant community-associated meticillin-resistant Staphylococcus aureus clone
Abstract
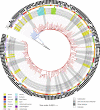
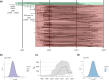
This study investigated the evolution and epidemiology of the community-associated and multidrug-resistant Staphylococcus aureus clone European CC1-MRSA-IV. Whole-genome sequences were obtained for 194 European CC1-MRSA-IV isolates (189 of human and 5 of animal origin) from 12 countries, and 10 meticillin-susceptible precursors (from North-Eastern Romania; all of human origin) of the clone. Phylogenetic analysis was performed using a maximum-likelihood approach, a time-measured phylogeny was reconstructed using Bayesian analysis, and in silico microarray genotyping was performed to identify resistance, virulence-associated and SCCmec (staphylococcal cassette chromosome mec) genes. Isolates were typically sequence type 1 (190/204) and spa type t127 (183/204). Bayesian analysis indicated that European CC1-MRSA-IV emerged in approximately 1995 before undergoing rapid expansion in the late 1990s and 2000s, while spreading throughout Europe and into the Middle East. Phylogenetic analysis revealed an unstructured meticillin-resistant S. aureus (MRSA) population, lacking significant geographical or temporal clusters. The MRSA were genotypically multidrug-resistant, consistently encoded seh, and intermittently (34/194) encoded an undisrupted hlb gene with concomitant absence of the lysogenic phage-encoded genes sak and scn. All MRSA also harboured a characteristic ~5350 nt insertion in SCCmec adjacent to orfX. Detailed demographic data from Denmark showed that there, the clone is typically (25/35) found in the community, and often (10/35) among individuals with links to South-Eastern Europe. This study elucidated the evolution and epidemiology of European CC1-MRSA-IV, which emerged from a meticillin-susceptible lineage prevalent in North-Eastern Romania before disseminating rapidly throughout Europe.
Keywords: CA-MRSA; European CC1-MRSA-IV clone; epidemiology; evolution; phylogenomics; transmission.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
References
-
- Carroll D, Kehoe MA, Cavanagh D, Coleman DC. Novel organization of the site-specific integration and excision recombination functions of the Staphylococcus aureus serotype F virulence-converting phages φ13 and φ42. Mol Microbiol. 1995;16:877–893. doi: 10.1111/j.1365-2958.1995.tb02315.x. - DOI - PubMed
-
- Monecke S, König E, Earls MR, Leitner E, Müller E, et al. An epidemic CC1-MRSA-IV clone yields false-negative test results in molecular MRSA identification assays: a note of caution, Austria, Germany, Ireland, 2020. Euro Surveill. 2020;25:pii=2000929. doi: 10.2807/1560-7917.ES.2020.25.25.2000929. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

