Resin-Assisted Capture Coupled with Isobaric Tandem Mass Tag Labeling for Multiplexed Quantification of Protein Thiol Oxidation
- PMID: 34223836
- PMCID: PMC8828046
- DOI: 10.3791/62671
Resin-Assisted Capture Coupled with Isobaric Tandem Mass Tag Labeling for Multiplexed Quantification of Protein Thiol Oxidation
Abstract
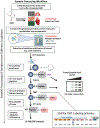
Reversible oxidative modifications on protein thiols have recently emerged as important mediators of cellular function. Herein we describe the detailed procedure of a quantitative redox proteomics method that utilizes resin-assisted capture (RAC) in combination with tandem mass tag (TMT) isobaric labeling and liquid chromatography-tandem mass spectrometry (LC-MS/MS) to allow multiplexed stochiometric quantification of oxidized protein thiols at the proteome level. The site-specific quantitative information on oxidized cysteine residues provides additional insight into the functional impacts of such modifications. The workflow is adaptable across many sample types, including cultured cells (e.g., mammalian, prokaryotic) and whole tissues (e.g., heart, lung, muscle), which are initially lysed/homogenized and with free thiols being alkylated to prevent artificial oxidation. The oxidized protein thiols are then reduced and captured by a thiol-affinity resin, which streamlines and simplifies the workflow steps by allowing the proceeding digestion, labeling, and washing procedures to be performed without additional transfer of proteins/peptides. Finally, the labeled peptides are eluted and analyzed by LC-MS/MS to reveal comprehensive stoichiometric changes related to thiol oxidation across the entire proteome. This method greatly improves the understanding of the role of redox-dependent regulation under physiological and pathophysiological states related to protein thiol oxidation.
Conflict of interest statement
Disclosures
The authors declare no conflicts of interest, financial or otherwise.
Figures

Similar articles
-
Resin-assisted enrichment of thiols as a general strategy for proteomic profiling of cysteine-based reversible modifications.Nat Protoc. 2014 Jan;9(1):64-75. doi: 10.1038/nprot.2013.161. Epub 2013 Dec 12. Nat Protoc. 2014. PMID: 24336471 Free PMC article.
-
Solid-phase capture for the detection and relative quantification of S-nitrosoproteins by mass spectrometry.Methods. 2013 Aug 1;62(2):130-7. doi: 10.1016/j.ymeth.2012.10.001. Epub 2012 Oct 11. Methods. 2013. PMID: 23064468 Free PMC article. Review.
-
Quantifying reversible oxidation of protein thiols in photosynthetic organisms.J Am Soc Mass Spectrom. 2015 Apr;26(4):631-40. doi: 10.1007/s13361-014-1073-y. Epub 2015 Feb 20. J Am Soc Mass Spectrom. 2015. PMID: 25698223
-
Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specific redox proteomics.Mol Cell Proteomics. 2014 Dec;13(12):3270-85. doi: 10.1074/mcp.M114.041160. Epub 2014 Aug 12. Mol Cell Proteomics. 2014. PMID: 25118246 Free PMC article.
-
Characterization of cellular oxidative stress response by stoichiometric redox proteomics.Am J Physiol Cell Physiol. 2021 Feb 1;320(2):C182-C194. doi: 10.1152/ajpcell.00040.2020. Epub 2020 Dec 2. Am J Physiol Cell Physiol. 2021. PMID: 33264075 Free PMC article. Review.
Cited by
-
A deep redox proteome profiling workflow and its application to skeletal muscle of a Duchenne Muscular Dystrophy model.Free Radic Biol Med. 2022 Nov 20;193(Pt 1):373-384. doi: 10.1016/j.freeradbiomed.2022.10.300. Epub 2022 Oct 25. Free Radic Biol Med. 2022. PMID: 36306991 Free PMC article.
-
Low-input redoxomics facilitates global identification of metabolic regulators of oxidative stress in the gut.Signal Transduct Target Ther. 2025 Jan 8;10(1):8. doi: 10.1038/s41392-024-02094-7. Signal Transduct Target Ther. 2025. PMID: 39774148 Free PMC article.
-
Integrating Redox Proteomics and Computational Modeling to Decipher Thiol-Based Oxidative Post-Translational Modifications (oxiPTMs) in Plant Stress Physiology.Int J Mol Sci. 2025 Jul 18;26(14):6925. doi: 10.3390/ijms26146925. Int J Mol Sci. 2025. PMID: 40725172 Free PMC article. Review.
-
Integrative Multi-PTM Proteomics Reveals Dynamic Global, Redox, Phosphorylation, and Acetylation Regulation in Cytokine-Treated Pancreatic Beta Cells.Mol Cell Proteomics. 2024 Dec;23(12):100881. doi: 10.1016/j.mcpro.2024.100881. Epub 2024 Nov 15. Mol Cell Proteomics. 2024. PMID: 39550035 Free PMC article.
-
Defining the S-Glutathionylation Proteome by Biochemical and Mass Spectrometric Approaches.Antioxidants (Basel). 2022 Nov 17;11(11):2272. doi: 10.3390/antiox11112272. Antioxidants (Basel). 2022. PMID: 36421458 Free PMC article. Review.
References
-
- Sies H, Jones DP Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nature Reviews Molecular Cell Biology. 21 (7), 363–383 (2020). - PubMed
-
- Olson KR The biological legacy of sulfur: A roadmap to the future. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 252, 110824 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
