Dynamic chromatin association of IκBα is regulated by acetylation and cleavage of histone H4
- PMID: 34224210
- PMCID: PMC8344903
- DOI: 10.15252/embr.202152649
Dynamic chromatin association of IκBα is regulated by acetylation and cleavage of histone H4
Abstract
IκBs exert principal functions as cytoplasmic inhibitors of NF-kB transcription factors. Additional roles for IκB homologues have been described, including chromatin association and transcriptional regulation. Phosphorylated and SUMOylated IκBα (pS-IκBα) binds to histones H2A and H4 in the stem cell and progenitor cell compartment of skin and intestine, but the mechanisms controlling its recruitment to chromatin are largely unknown. Here, we show that serine 32-36 phosphorylation of IκBα favors its binding to nucleosomes and demonstrate that p-IκBα association with H4 depends on the acetylation of specific H4 lysine residues. The N-terminal tail of H4 is removed during intestinal cell differentiation by proteolytic cleavage by trypsin or chymotrypsin at residues 17-19, which reduces p-IκBα binding. Inhibition of trypsin and chymotrypsin activity in HT29 cells increases p-IκBα chromatin binding but, paradoxically, impaired goblet cell differentiation, comparable to IκBα deletion. Taken together, our results indicate that dynamic binding of IκBα to chromatin is a requirement for intestinal cell differentiation and provide a molecular basis for the understanding of the restricted nuclear distribution of p-IκBα in specific stem cell compartments.
Keywords: differentiation; histone H4; histone cleavage; intestine; nuclear IkappaB.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

PD experiments under different salt concentrations using GST‐H4 as bait and the indicated IκBα constructs expressed in HEK‐293T cells.
Coomassie staining analysis of the indicated fractions recovered in the Fast Protein Liquid Chromatography (FPLC) analysis of IκBα and histone H2A complexes.
Western blot analysis of IκBα phosphorylated in vitro by addition of active IKKβ kinase with anti‐p‐IκBα (S32‐36) antibody.
PD experiment using GST‐H4 as bait and the indicated IκBα mutants. Quantification of the interaction from 3 biological replicates performed. Bars represent mean values ± standard deviation of the replicates. Statistical analysis of different was obtained by t‐test; *P < 0.05, **P < 0.01.

Electrophoretic analysis (under non‐denaturing conditions) of the association between the indicated IκBα species association and reconstituted nucleosome core particles (NCP).
Electrophoretic analysis in agarose gels of in vitro generated p‐IκBα and reconstituted nucleosome in absence or presence of anti‐IκBα antibodies.

PD experiment with biotinylated peptides of H4 (1–23aa) and total lysates of HCT‐116 cells.
Quantification of the IκBα 60 kDa band (upper panels) and the 37 kDa band (lower panels) is shown as average and s.d. of three independent experiments.
PD experiments with GST‐SUMO IκBα fusion protein and serial dilutions of histone‐enriched HEK‐293T lysates. Quantification of three independent replicates is shown.
Venn diagrams representing the overlap between IκBα and acetylated H4 peaks obtained in ChIP‐seq experiments from HCT‐116 cells.
Peak distribution of acetylated H4, H4K12ac and total histone H4 obtained in IκBα target genes and non‐IκBα targets relative to the transcription start site (TSS).
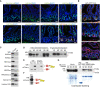
Immunofluorescence (IF) analysis with the indicated antibodies in sections from murine small intestine of 2‐month‐old WT mice.
Double IF analysis with the indicated antibodies in sections from murine small intestine of 2‐month‐old Lgr5‐GFP transgenic mice.
Western blot analysis of chromatin extracts from isolated intestinal villus and crypt cells.
Western blot analysis of chromatin extracts from isolated intestinal villus and crypt cells in several 2‐month‐old WT mice (each # represents a mouse).
Western blot analysis of soluble (Sol) and chromatin (Chr) extracts from isolated intestinal villus and crypt cells in 2‐month‐old mCherry‐H4 transgenic mice.
Western blot analysis of chromatin fraction from mCherry‐H4 transfected HCT‐116 cells.
Pull‐down assay using GST‐SUMO IκBα protein and chromatin lysates from 2‐month‐old WT mice villi.

IF analysis with the indicated antibodies in sections from skin of 2‐month‐old mice.
IF analysis of human colonic tissue with the indicated antibodies.
IF analysis of sections from small intestine of mice at different stages of development.

- A
Western blot analysis of the crypt‐derived chromatin fractions incubated with soluble lysates from villi or crypts for the indicated times.
- B
Table showing the proteases differentially expressed in the villus compartment as determined by RNA‐seq analysis of intestinal crypt (purified EphB2 high cells) and villus (EphB2 negative/low) cells. Predicted cleavage sites for each protease in the histone H4 sequence were determined using the PeptideCutter‐ExPASy and PROSPER software.
- C
Western blot analysis of mCherry‐H4 with the indicated mutations transfected in HCT‐116 cells. Quantification of relative amounts of truncated H4 relative to the WT.
- D, E
Western blot analysis of mCherry‐H4 transfected in HCT‐116 cells treated for 16 hours with TLCK (D) or AdaAhx₃L₃VS (E) at the indicated concentrations.
- F
Western blot analysis of a cleavage experiment incubating modified H4 peptides (P1‐P9) with soluble lysates from villi or crypts in the presence (+) or absence (−) of the commercial protease inhibitors cocktail. Lanes indicated as P correspond to the control peptide without lysate incubation. Notice the electrophoretic shift of peptides incubated with villus lysates (compared with control peptides) suggestive of post‐translational modifications or binding to proteins absent from crypt lysates.
- G
Quantification of the relative cleavage of specific H4 peptides incubated with villus‐derived lysates from 3 independent experiments performed.
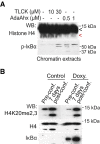
WB analysis of chromatin extracts from HT29 cells treated as indicated. The red arrowhead indicates truncated histone H4.
WB analysis of chromatin extracts from IκBα KO HT29 cells carrying doxycycline‐inducible IκBα expression construct untreated or treated with doxycycline for 16 h.

- A
Western blot analysis of soluble and chromatin extracts from HT29 cells at different differentiation stages (days post‐confluence).
- B, C
qPCR (B) and WB analysis (C) of the indicated genes or proteins in parental or IκBα KO HT29 cells obtained at pre‐confluence or 7‐day post‐confluence. Bars represent mean values ± standard deviation of 3 technical replicates from 3 biological replicates performed.
- D, E
qPCR (D) and WB analysis (E) of MUC5AC in HT29 cells treated with the indicated protease inhibitors. Bars represent mean values ± standard deviation from 3 biological replicates performed.
- F, G
qPCR (F) and WB analysis (G) of MUC5AC in HT29 cells transduced with sh‐RNAs against trypsin (T1, T2) or chymotrypsin (C1, C2). Bars represent mean values ± standard deviation of 3 technical replicates from 3 biological replicates performed.
- H
Western blot analysis of cells transfected with mCherry‐H4 and the indicated sh‐RNAs.
- I
Model for gene regulation by chromatin‐associated IκBα. In brief, dynamic dissociation of IκBα from the chromatin at specific genetic loci promotes transcription of genes involved in stem cell maturation or differentiation (upper panel). Absence of dynamic binding (middle panel) or cells IκBα deficient (lower panels) fail to activate IκBα‐dependent transcription thus imposing a differentiation/maturation blockage.
- A, B
Analysis by qPCR of SPDEF in HT29 cells WT or IκBα KO (A) or transduced with specific shRNA against trypsin (T) or chymotrypsin (C) at pre‐confluence or 7 days after confluence (B). Bars represent mean values ± standard deviation from 3 biological replicates performed.
- C
qPCR analysis of the muco‐secretory differentiation marker MUC2 in mouse intestinal organoids treated as indicated. Bars represent mean values ± standard deviation from 3 biological replicates performed.
- D
qPCR analysis of the stem cell marker Lgr5 in HT29 cells treated with the indicated inhibitors. Bars represent mean values ± standard deviation from 3 biological replicates performed.
Comment in
-
Histone clipping: the punctuation in the histone code.EMBO Rep. 2021 Aug 4;22(8):e53440. doi: 10.15252/embr.202153440. Epub 2021 Jul 7. EMBO Rep. 2021. PMID: 34232560 Free PMC article.
References
-
- Arenzana‐Seisdedos F, Turpin P, Rodriguez M, Thomas D, Hay RT, Virelizier JL, Dargemont C (1997) Nuclear localization of IκBα promotes active transport of NF‐κB from the nucleus to the cytoplasm. J Cell Sci 110: 369–378 - PubMed
-
- Azad GK, Swagatika S, Kumawat M, Kumawat R, Tomar RS (2018) Modifying chromatin by histone tail clipping. J Mol Biol 430: 3051–3067 - PubMed
-
- de Bolos C , Real FX, Lopez‐Ferrer A (2001) Regulation of mucin and glycoconjugate expression: from normal epithelium to gastric tumors. Front Biosci 6: 1256–1263 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

