The integration of differentially expressed genes based on multiple microarray datasets for prediction of the prognosis in oral squamous cell carcinoma
- PMID: 34224327
- PMCID: PMC8806768
- DOI: 10.1080/21655979.2021.1947076
The integration of differentially expressed genes based on multiple microarray datasets for prediction of the prognosis in oral squamous cell carcinoma
Abstract
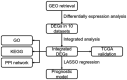
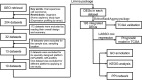
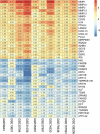
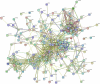
Oral squamous cell carcinoma (OSCC) is a common human malignancy. However, its pathogenesis and prognostic information are poorly elucidated. In the present study, we aimed to probe the most significant differentially expressed genes (DEGs) and their prognostic performance in OSCC. Multiple microarray datasets from the Gene Expression Omnibus (GEO) database were aggregated to identify DEGs between OSCC tissue and control tissue. Least absolute shrinkage and selection operator (LASSO) Cox model was constructed to determine the prognostic performance of the aggregated DEGs based on The Cancer Genome Atlas (TCGA) OSCC cohort. Ten datasets with 341 OSCC samples and 283 control samples were included. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment revealed that the integrated DEGs were enriched in the IL-17 signaling pathway, viral protein interactions with cytokines and cytokine receptors, and amoebiasis, among others. Our LASSO Cox model was able to discriminate two groups with different overall survival in the training cohort and test cohort (p < 0.001). The time-dependent receiver operating characteristic (ROC) curve revealed that the area under the curve (AUC) values at one year, three years, and five years were 0.831, 0.898, and 0.887, respectively. In the testing cohort, the time-dependent ROC curve showed that the AUC values at one year, three years, and five years were 0.696, 0.693, and 0.860, respectively. Our study showed that the integrated DEGs of OSCC might be applicable in the evaluation of prognosis in OSCC. However, further research should be performed to validate our findings.
Keywords: GEO; Oral squamous cell carcinoma; TCGA; bioinformatics; biomarker; prognosis.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




Similar articles
-
A Five-mRNA Expression Signature to Predict Survival in Oral Squamous Cell Carcinoma by Integrated Bioinformatic Analyses.Genet Test Mol Biomarkers. 2021 Aug;25(8):517-527. doi: 10.1089/gtmb.2021.0066. Genet Test Mol Biomarkers. 2021. PMID: 34406843 Free PMC article.
-
Identification for Exploring Underlying Pathogenesis and Therapy Strategy of Oral Squamous Cell Carcinoma by Bioinformatics Analysis.Med Sci Monit. 2019 Dec 3;25:9216-9226. doi: 10.12659/MSM.917736. Med Sci Monit. 2019. PMID: 31794546 Free PMC article.
-
Exploring Renin-angiotensin System Genes as Novel Prognostic Biomarkers for Oral Squamous Cell Carcinoma.Int J Med Sci. 2025 Apr 28;22(10):2470-2487. doi: 10.7150/ijms.112735. eCollection 2025. Int J Med Sci. 2025. PMID: 40386053 Free PMC article.
-
Identification of a Gene Prognostic Signature for Oral Squamous Cell Carcinoma by RNA Sequencing and Bioinformatics.Biomed Res Int. 2021 Apr 1;2021:6657767. doi: 10.1155/2021/6657767. eCollection 2021. Biomed Res Int. 2021. PMID: 33869632 Free PMC article.
-
A Narrative Review of Prognostic Gene Signatures in Oral Squamous Cell Carcinoma Using LASSO Cox Regression.Biomedicines. 2025 Jan 8;13(1):134. doi: 10.3390/biomedicines13010134. Biomedicines. 2025. PMID: 39857718 Free PMC article. Review.
Cited by
-
Potential Biomarkers and Endometrial Immune Microenvironment in Recurrent Implantation Failure.Biomolecules. 2023 Feb 21;13(3):406. doi: 10.3390/biom13030406. Biomolecules. 2023. PMID: 36979341 Free PMC article.
-
Long non-coding RNA CASC9 promotes tumor progression in oral squamous cell carcinoma by regulating microRNA-545-3p/laminin subunit gamma 2.Bioengineered. 2021 Dec;12(1):7907-7919. doi: 10.1080/21655979.2021.1977103. Bioengineered. 2021. PMID: 34612783 Free PMC article.
-
The Role of IL-17 in the Pathogenesis of Oral Squamous Cell Carcinoma.Int J Mol Sci. 2023 Jun 8;24(12):9874. doi: 10.3390/ijms24129874. Int J Mol Sci. 2023. PMID: 37373022 Free PMC article. Review.
-
SKA1 promotes oncogenic properties in oral dysplasia and oral squamous cell carcinoma, and augments resistance to radiotherapy.Mol Oncol. 2025 Apr;19(4):1054-1074. doi: 10.1002/1878-0261.13780. Epub 2024 Dec 10. Mol Oncol. 2025. PMID: 39656562 Free PMC article.
-
MicroRNA-26b-5p suppresses the proliferation of tongue squamous cell carcinoma via targeting proline rich 11 (PRR11).Bioengineered. 2021 Dec;12(1):5830-5838. doi: 10.1080/21655979.2021.1969832. Bioengineered. 2021. PMID: 34488538 Free PMC article.
References
-
- Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BIOCONDUCTOR. Bioinformatics. 2007;23(14):1846–1847. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
